Architecture and DNA recognition elements of the Fanconi anemia FANCM-FAAP24 complex
- PMID: 23932590
- PMCID: PMC3763369
- DOI: 10.1016/j.str.2013.07.006
Architecture and DNA recognition elements of the Fanconi anemia FANCM-FAAP24 complex
Abstract
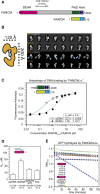
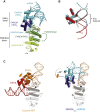
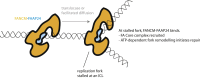
Fanconi anemia (FA) is a disorder associated with a failure in DNA repair. FANCM (defective in FA complementation group M) and its partner FAAP24 target other FA proteins to sites of DNA damage. FANCM-FAAP24 is related to XPF/MUS81 endonucleases but lacks endonucleolytic activity. We report a structure of an FANCM C-terminal fragment (FANCMCTD) bound to FAAP24 and DNA. This S-shaped structure reveals the FANCM (HhH)2 domain is buried, whereas the FAAP24 (HhH)2 domain engages DNA. We identify a second DNA contact and a metal center within the FANCM pseudo-nuclease domain and demonstrate that mutations in either region impair double-stranded DNA binding in vitro and FANCM-FAAP24 function in vivo. We show the FANCM translocase domain lies in proximity to FANCMCTD by electron microscopy and that binding fork DNA structures stimulate its ATPase activity. This suggests a tracking model for FANCM-FAAP24 until an encounter with a stalled replication fork triggers ATPase-mediated fork remodeling.
Copyright © 2013 Elsevier Ltd. All rights reserved.
Figures


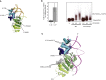
Similar articles
-
Structural insights into the functions of the FANCM-FAAP24 complex in DNA repair.Nucleic Acids Res. 2013 Dec;41(22):10573-83. doi: 10.1093/nar/gkt788. Epub 2013 Sep 3. Nucleic Acids Res. 2013. PMID: 24003026 Free PMC article.
-
Identification of FAAP24, a Fanconi anemia core complex protein that interacts with FANCM.Mol Cell. 2007 Feb 9;25(3):331-43. doi: 10.1016/j.molcel.2007.01.003. Mol Cell. 2007. PMID: 17289582
-
Structure analysis of FAAP24 reveals single-stranded DNA-binding activity and domain functions in DNA damage response.Cell Res. 2013 Oct;23(10):1215-28. doi: 10.1038/cr.2013.124. Epub 2013 Sep 3. Cell Res. 2013. PMID: 23999858 Free PMC article.
-
FANCM-FAAP24 and FANCJ: FA proteins that metabolize DNA.Mutat Res. 2009 Jul 31;668(1-2):20-6. doi: 10.1016/j.mrfmmm.2009.04.002. Epub 2009 Apr 18. Mutat Res. 2009. PMID: 19379763 Free PMC article. Review.
-
FANCM-FAAP24 and HCLK2: roles in ATR signalling and the Fanconi anemia pathway.Cell Cycle. 2009 Apr 15;8(8):1133-7. doi: 10.4161/cc.8.8.8204. Epub 2009 Apr 16. Cell Cycle. 2009. PMID: 19282663 Review.
Cited by
-
The Fanconi Anemia Pathway Maintains Genome Stability by Coordinating Replication and Transcription.Mol Cell. 2015 Nov 5;60(3):351-61. doi: 10.1016/j.molcel.2015.09.012. Epub 2015 Oct 22. Mol Cell. 2015. PMID: 26593718 Free PMC article.
-
Mechanism of structure-specific DNA binding by the FANCM branchpoint translocase.Nucleic Acids Res. 2024 Oct 14;52(18):11029-11044. doi: 10.1093/nar/gkae727. Nucleic Acids Res. 2024. PMID: 39189453 Free PMC article.
-
The FA Core Complex Contains a Homo-dimeric Catalytic Module for the Symmetric Mono-ubiquitination of FANCI-FANCD2.Cell Rep. 2017 Jan 17;18(3):611-623. doi: 10.1016/j.celrep.2016.11.013. Epub 2016 Dec 13. Cell Rep. 2017. PMID: 27986592 Free PMC article.
-
Recent discoveries in the molecular pathogenesis of the inherited bone marrow failure syndrome Fanconi anemia.Blood Rev. 2017 May;31(3):93-99. doi: 10.1016/j.blre.2016.10.002. Epub 2016 Oct 13. Blood Rev. 2017. PMID: 27760710 Free PMC article. Review.
-
ALTernative Functions for Human FANCM at Telomeres.Front Mol Biosci. 2019 Sep 6;6:84. doi: 10.3389/fmolb.2019.00084. eCollection 2019. Front Mol Biosci. 2019. PMID: 31552268 Free PMC article. Review.
References
-
- Berger I., Fitzgerald D.J., Richmond T.J. Baculovirus expression system for heterologous multiprotein complexes. Nat. Biotechnol. 2004;22:1583–1587. - PubMed
-
- Blackford A.N., Schwab R.A., Nieminuszczy J., Deans A.J., West S.C., Niedzwiedz W. The DNA translocase activity of FANCM protects stalled replication forks. Hum. Mol. Genet. 2012;21:2005–2016. - PubMed
-
- Ciccia A., Ling C., Coulthard R., Yan Z., Xue Y., Meetei A.R., Laghmani H., Joenje H., McDonald N., de Winter J.P. Identification of FAAP24, a Fanconi anemia core complex protein that interacts with FANCM. Mol. Cell. 2007;25:331–343. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

