The DNA helicase-primase complex as a target for herpes viral infection
- PMID: 23930666
- PMCID: PMC4098783
- DOI: 10.1517/14728222.2013.827663
The DNA helicase-primase complex as a target for herpes viral infection
Abstract
Introduction: The Herpesviridae are responsible for debilitating acute and chronic infections, and some members of this family are associated with human cancers. Conventional anti-herpesviral therapy targets the viral DNA polymerase and has been extremely successful; however, the emergence of drug-resistant virus strains, especially in neonates and immunocompromised patients, underscores the need for continued development of anti-herpes drugs. In this article, we explore an alternative target for antiviral therapy, the HSV helicase/primase complex.
Areas covered: This review addresses the current state of knowledge of HSV DNA replication and the important roles played by the herpesvirus helicase- primase complex. In the last 10 years several helicase/primase inhibitors (HPIs) have been described, and in this article, we discuss and contrast these new agents with established inhibitors.
Expert opinion: The outstanding safety profile of existing nucleoside analogues for α-herpesvirus infection make the development of new therapeutic agents a challenge. Currently used nucleoside analogues exhibit few side effects and have low occurrence of clinically relevant resistance. For HCMV, however, existing drugs have significant toxicity issues and the frequency of drug resistance is high, and no antiviral therapies are available for EBV and KSHV. The development of new anti-herpesvirus drugs is thus well worth pursuing especially for immunocompromised patients and those who develop drug-resistant infections. Although the HPIs are promising, limitations to their development into a successful drug strategy remain.
Conflict of interest statement
The authors state no conflict of interest and have received no payment in preparation of this manuscript.
Figures

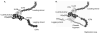
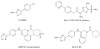
Similar articles
-
A short commentary on the review by Weller and Kuchta: the DNA helicase-primase complex as a target for herpes virus infection.Expert Opin Ther Targets. 2013 Oct;17(10):1113-4. doi: 10.1517/14728222.2013.840294. Expert Opin Ther Targets. 2013. PMID: 24044544 No abstract available.
-
High conservation of herpes simplex virus UL5/UL52 helicase-primase complex in the era of new antiviral therapies.Antiviral Res. 2016 Apr;128:1-6. doi: 10.1016/j.antiviral.2016.01.015. Epub 2016 Jan 28. Antiviral Res. 2016. PMID: 26826441
-
Herpes simplex virus helicase-primase inhibitors: recent findings from the study of drug resistance mutations.Antivir Chem Chemother. 2008;19(1):1-6. doi: 10.1177/095632020801900101. Antivir Chem Chemother. 2008. PMID: 18610552 Review.
-
Helicase primase inhibitors (HPIs) are efficacious for therapy of human herpes simplex virus (HSV) disease in an infection mouse model.Antiviral Res. 2021 Nov;195:105190. doi: 10.1016/j.antiviral.2021.105190. Epub 2021 Oct 16. Antiviral Res. 2021. PMID: 34666109
-
Antiviral drug resistance and helicase-primase inhibitors of herpes simplex virus.Drug Resist Updat. 2011 Feb;14(1):45-51. doi: 10.1016/j.drup.2010.11.002. Epub 2010 Dec 22. Drug Resist Updat. 2011. PMID: 21183396 Review.
Cited by
-
Coronavirus helicases: attractive and unique targets of antiviral drug-development and therapeutic patents.Expert Opin Ther Pat. 2021 Apr;31(4):339-350. doi: 10.1080/13543776.2021.1884224. Epub 2021 Apr 21. Expert Opin Ther Pat. 2021. PMID: 33593200 Free PMC article. Review.
-
Current Perspectives on the Management of Herpesvirus Infections in Solid Organ Transplant Recipients.Viruses. 2023 Jul 21;15(7):1595. doi: 10.3390/v15071595. Viruses. 2023. PMID: 37515280 Free PMC article. Review.
-
Antimicrobial and antiviral evaluation of compounds from Holoptelea integrifolia: in silico supported in vitro study.RSC Adv. 2023 Nov 3;13(46):32473-32486. doi: 10.1039/d3ra05978b. eCollection 2023 Oct 31. RSC Adv. 2023. PMID: 37928846 Free PMC article.
-
Full Genome Sequence-Based Comparative Study of Wild-Type and Vaccine Strains of Infectious Laryngotracheitis Virus from Italy.PLoS One. 2016 Feb 18;11(2):e0149529. doi: 10.1371/journal.pone.0149529. eCollection 2016. PLoS One. 2016. PMID: 26890525 Free PMC article.
-
UL52 primase interactions in the herpes simplex virus 1 helicase-primase are affected by antiviral compounds and mutations causing drug resistance.J Biol Chem. 2014 Nov 21;289(47):32583-92. doi: 10.1074/jbc.M114.609453. Epub 2014 Oct 2. J Biol Chem. 2014. PMID: 25278021 Free PMC article.
References
-
- Chayavichitsilp P, Buckwalter JV, Krakowski AC, Friedlander SF. Herpes simplex. Ped Rev. 2009;30:119–29. - PubMed
-
- Kimberlin DW, Whitley RJ. Antiviral therapy of HSV-1 and -2. In: Arvin A, Campadelli-Fiume G, Mocarski E, et al., editors. Human herpesviruses: biology, therapy, and immunoprophylaxis. Cambridge University Press; Cambridge: 2007. - PubMed
-
- Crumpacker CS. Ganciclovir. N Engl J Med. 1996;335:721–9. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
