REEP3/4 ensure endoplasmic reticulum clearance from metaphase chromatin and proper nuclear envelope architecture
- PMID: 23911198
- PMCID: PMC3745822
- DOI: 10.1016/j.devcel.2013.06.016
REEP3/4 ensure endoplasmic reticulum clearance from metaphase chromatin and proper nuclear envelope architecture
Abstract
Dynamic interactions between membrane-bound organelles and the microtubule cytoskeleton are crucial to establish, maintain, and remodel the internal organization of cells throughout the cell cycle. However, the molecular nature of these interactions remains poorly understood. We performed a biochemical screen for microtubule-membrane linkers and identified REEP4, a previously uncharacterized endoplasmic reticulum (ER) protein. Depletion of REEP4 and the closely related REEP3 from HeLa cells causes defects in cell division and a proliferation of intranuclear membranes derived from the nuclear envelope. This phenotype originates in mitosis, when ER membranes accumulate on metaphase chromosomes. Microtubule binding and mitotic ER clearance from chromosomes both depend on a short, positively charged amino acid sequence connecting the two hydrophobic domains of REEP4. Our results show that REEP3/4 function redundantly to clear the ER from metaphase chromatin, thereby ensuring correct progression through mitosis and proper nuclear envelope architecture.
Copyright © 2013 Elsevier Inc. All rights reserved.
Figures
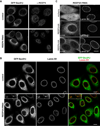
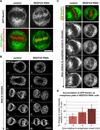
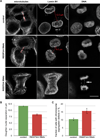
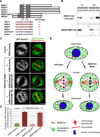
Comment in
-
PREEParing for mitosis.Dev Cell. 2013 Aug 12;26(3):221-2. doi: 10.1016/j.devcel.2013.07.018. Dev Cell. 2013. PMID: 23948250
Similar articles
-
REEP3 and REEP4 determine the tubular morphology of the endoplasmic reticulum during mitosis.Mol Biol Cell. 2019 Jun 1;30(12):1377-1389. doi: 10.1091/mbc.E18-11-0698. Epub 2019 Apr 17. Mol Biol Cell. 2019. PMID: 30995177 Free PMC article.
-
PREEParing for mitosis.Dev Cell. 2013 Aug 12;26(3):221-2. doi: 10.1016/j.devcel.2013.07.018. Dev Cell. 2013. PMID: 23948250
-
Microtubules as key coordinators of nuclear envelope and endoplasmic reticulum dynamics during mitosis.Bioessays. 2014 Jul;36(7):665-71. doi: 10.1002/bies.201400022. Epub 2014 May 21. Bioessays. 2014. PMID: 24848719 Review.
-
Dissociation of membrane-chromatin contacts is required for proper chromosome segregation in mitosis.Mol Biol Cell. 2019 Feb 15;30(4):427-440. doi: 10.1091/mbc.E18-10-0609. Epub 2018 Dec 26. Mol Biol Cell. 2019. PMID: 30586323 Free PMC article.
-
Sorting out the nuclear envelope from the endoplasmic reticulum.Nat Rev Mol Cell Biol. 2004 Jan;5(1):65-9. doi: 10.1038/nrm1263. Nat Rev Mol Cell Biol. 2004. PMID: 14663490 Review.
Cited by
-
Endoplasmic reticulum membranes are continuously required to maintain mitotic spindle size and forces.Life Sci Alliance. 2022 Nov 15;6(1):e202201540. doi: 10.26508/lsa.202201540. Print 2023 Jan. Life Sci Alliance. 2022. PMID: 36379670 Free PMC article.
-
Microtubules are necessary for proper Reticulon localization during mitosis.PLoS One. 2019 Dec 26;14(12):e0226327. doi: 10.1371/journal.pone.0226327. eCollection 2019. PLoS One. 2019. PMID: 31877164 Free PMC article.
-
Organelle size scaling over embryonic development.Wiley Interdiscip Rev Dev Biol. 2020 Sep;9(5):e376. doi: 10.1002/wdev.376. Epub 2020 Jan 31. Wiley Interdiscip Rev Dev Biol. 2020. PMID: 32003549 Free PMC article. Review.
-
The Phytochemical Rhein Mediates M6A-Independent Suppression of Adipocyte Differentiation.Front Nutr. 2021 Nov 1;8:756803. doi: 10.3389/fnut.2021.756803. eCollection 2021. Front Nutr. 2021. PMID: 34790688 Free PMC article.
-
REEP3 and REEP4 determine the tubular morphology of the endoplasmic reticulum during mitosis.Mol Biol Cell. 2019 Jun 1;30(12):1377-1389. doi: 10.1091/mbc.E18-11-0698. Epub 2019 Apr 17. Mol Biol Cell. 2019. PMID: 30995177 Free PMC article.
References
-
- Beaudouin J, Gerlich D, Daigle N, Eils R, Ellenberg J. Nuclear envelope breakdown proceeds by microtubule-induced tearing of the lamina. Cell. 2002;108:83–96. - PubMed
-
- Foisner R, Gerace L. Integral membrane proteins of the nuclear envelope interact with lamins and chromosomes, and binding is modulated by mitotic phosphorylation. Cell. 1993;73:1267–1279. - PubMed
-
- Hannak E, Heald R. Investigating mitotic spindle assembly and function using Xenopus laevis egg extracts. Nat. Protoc. 2006;1:2305–2314. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

