Metformin improves healthspan and lifespan in mice
- PMID: 23900241
- PMCID: PMC3736576
- DOI: 10.1038/ncomms3192
Metformin improves healthspan and lifespan in mice
Abstract
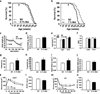
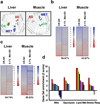
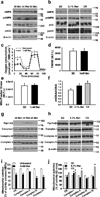
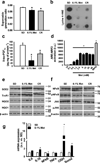
Metformin is a drug commonly prescribed to treat patients with type 2 diabetes. Here we show that long-term treatment with metformin (0.1% w/w in diet) starting at middle age extends healthspan and lifespan in male mice, while a higher dose (1% w/w) was toxic. Treatment with metformin mimics some of the benefits of calorie restriction, such as improved physical performance, increased insulin sensitivity, and reduced low-density lipoprotein and cholesterol levels without a decrease in caloric intake. At a molecular level, metformin increases AMP-activated protein kinase activity and increases antioxidant protection, resulting in reductions in both oxidative damage accumulation and chronic inflammation. Our results indicate that these actions may contribute to the beneficial effects of metformin on healthspan and lifespan. These findings are in agreement with current epidemiological data and raise the possibility of metformin-based interventions to promote healthy aging.
Figures
Similar articles
-
A Critical Review of the Evidence That Metformin Is a Putative Anti-Aging Drug That Enhances Healthspan and Extends Lifespan.Front Endocrinol (Lausanne). 2021 Aug 5;12:718942. doi: 10.3389/fendo.2021.718942. eCollection 2021. Front Endocrinol (Lausanne). 2021. PMID: 34421827 Free PMC article. Review.
-
Metformin induces a dietary restriction-like state and the oxidative stress response to extend C. elegans Healthspan via AMPK, LKB1, and SKN-1.PLoS One. 2010 Jan 18;5(1):e8758. doi: 10.1371/journal.pone.0008758. PLoS One. 2010. PMID: 20090912 Free PMC article.
-
The Use of Metformin to Increase the Human Healthspan.Adv Exp Med Biol. 2020;1260:319-332. doi: 10.1007/978-3-030-42667-5_13. Adv Exp Med Biol. 2020. PMID: 32304040 Review.
-
Metformin and caloric restriction induce an AMPK-dependent restoration of mitochondrial dysfunction in fibroblasts from Fibromyalgia patients.Biochim Biophys Acta. 2015 Jul;1852(7):1257-67. doi: 10.1016/j.bbadis.2015.03.005. Epub 2015 Mar 14. Biochim Biophys Acta. 2015. PMID: 25779083
-
Metformin treatment of diverse Caenorhabditis species reveals the importance of genetic background in longevity and healthspan extension outcomes.Aging Cell. 2022 Jan;21(1):e13488. doi: 10.1111/acel.13488. Epub 2021 Nov 27. Aging Cell. 2022. PMID: 34837316 Free PMC article.
Cited by
-
Metformin activates AMP-activated protein kinase by promoting formation of the αβγ heterotrimeric complex.J Biol Chem. 2015 Feb 6;290(6):3793-802. doi: 10.1074/jbc.M114.604421. Epub 2014 Dec 23. J Biol Chem. 2015. PMID: 25538235 Free PMC article.
-
Nicandra physalodes Extract Exerts Antiaging Effects in Multiple Models and Extends the Lifespan of Caenorhabditis elegans via DAF-16 and HSF-1.Oxid Med Cell Longev. 2022 Oct 11;2022:3151071. doi: 10.1155/2022/3151071. eCollection 2022. Oxid Med Cell Longev. 2022. PMID: 36267808 Free PMC article.
-
L-threonine promotes healthspan by expediting ferritin-dependent ferroptosis inhibition in C. elegans.Nat Commun. 2022 Nov 2;13(1):6554. doi: 10.1038/s41467-022-34265-x. Nat Commun. 2022. PMID: 36323683 Free PMC article.
-
Urinary markers of oxidative stress respond to infection and late-life in wild chimpanzees.PLoS One. 2020 Sep 11;15(9):e0238066. doi: 10.1371/journal.pone.0238066. eCollection 2020. PLoS One. 2020. PMID: 32916689 Free PMC article.
-
Adverse Geriatric Outcomes Secondary to Polypharmacy in a Mouse Model: The Influence of Aging.J Gerontol A Biol Sci Med Sci. 2016 May;71(5):571-7. doi: 10.1093/gerona/glv046. Epub 2015 May 4. J Gerontol A Biol Sci Med Sci. 2016. PMID: 25940962 Free PMC article.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

