Clade-specific virulence patterns of Mycobacterium tuberculosis complex strains in human primary macrophages and aerogenically infected mice
- PMID: 23900170
- PMCID: PMC3735190
- DOI: 10.1128/mBio.00250-13
Clade-specific virulence patterns of Mycobacterium tuberculosis complex strains in human primary macrophages and aerogenically infected mice
Abstract
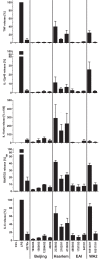
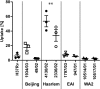
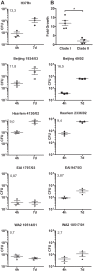
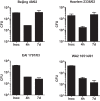
In infection experiments with genetically distinct Mycobacterium tuberculosis complex (MTBC) strains, we identified clade-specific virulence patterns in human primary macrophages and in mice infected by the aerosol route, both reflecting relevant model systems. Exclusively human-adapted M. tuberculosis lineages, also termed clade I, comprising "modern" lineages, such as Beijing and Euro-American Haarlem strains, showed a significantly enhanced capability to grow compared to that of clade II strains, which include "ancient" lineages, such as, e.g., East African Indian or M. africanum strains. However, a simple correlation of inflammatory response profiles with strain virulence was not apparent. Overall, our data reveal three different pathogenic profiles: (i) strains of the Beijing lineage are characterized by low uptake, low cytokine induction, and a high replicative potential, (ii) strains of the Haarlem lineage by high uptake, high cytokine induction, and high growth rates, and (iii) EAI strains by low uptake, low cytokine induction, and a low replicative potential. Our findings have significant implications for our understanding of host-pathogen interaction and factors that modulate the outcomes of infections. Future studies addressing the underlying mechanisms and clinical implications need to take into account the diversity of both the pathogen and the host.
Importance: Clinical strains of the Mycobacterium tuberculosis complex (MTBC) are genetically more diverse than previously anticipated. Our analysis of mycobacterial growth characteristics in primary human macrophages and aerogenically infected mice shows that the MTBC genetic differences translate into pathogenic differences in the interaction with the host. Our study reveals for the first time that "TB is not TB," if put in plain terms. We are convinced that it is very unlikely that a single molecular mechanism may explain the observed effects. Our study refutes the hypothesis that there is a simple correlation between cytokine induction as a single functional parameter of host interaction and mycobacterial virulence. Instead, careful consideration of strain- and lineage-specific characteristics must guide our attempts to decipher what determines the pathological potential and thus the outcomes of infection with MTBC, one of the most important human pathogens.
Figures


Similar articles
-
Shaping the niche in macrophages: Genetic diversity of the M. tuberculosis complex and its consequences for the infected host.Int J Med Microbiol. 2018 Jan;308(1):118-128. doi: 10.1016/j.ijmm.2017.09.009. Epub 2017 Sep 14. Int J Med Microbiol. 2018. PMID: 28969988 Review.
-
Consequences of genomic diversity in Mycobacterium tuberculosis.Semin Immunol. 2014 Dec;26(6):431-44. doi: 10.1016/j.smim.2014.09.012. Epub 2014 Oct 22. Semin Immunol. 2014. PMID: 25453224 Free PMC article. Review.
-
Mycobacterium tuberculosis strains of the modern sublineage of the Beijing family are more likely to display increased virulence than strains of the ancient sublineage.J Clin Microbiol. 2014 Jul;52(7):2615-24. doi: 10.1128/JCM.00498-14. Epub 2014 May 14. J Clin Microbiol. 2014. PMID: 24829250 Free PMC article.
-
Clinical isolates of the modern Mycobacterium tuberculosis lineage 4 evade host defense in human macrophages through eluding IL-1β-induced autophagy.Cell Death Dis. 2018 May 24;9(6):624. doi: 10.1038/s41419-018-0640-8. Cell Death Dis. 2018. PMID: 29795378 Free PMC article.
-
The Bioinformatics Analysis of Comparative Genomics of Mycobacterium tuberculosis Complex (MTBC) Provides Insight into Dissimilarities between Intraspecific Groups Differing in Host Association, Virulence, and Epitope Diversity.Front Cell Infect Microbiol. 2017 Mar 21;7:88. doi: 10.3389/fcimb.2017.00088. eCollection 2017. Front Cell Infect Microbiol. 2017. PMID: 28377903 Free PMC article.
Cited by
-
Draft genome sequence of Mycobacterium tuberculosis strain B9741 of Beijing B0/W lineage from HIV positive patient from Siberia.Genom Data. 2016 Aug 3;10:61-62. doi: 10.1016/j.gdata.2016.08.001. eCollection 2016 Dec. Genom Data. 2016. PMID: 27761405 Free PMC article.
-
Characterisation of secretome-based immune responses of human leukocytes infected with various Mycobacterium tuberculosis lineages.PeerJ. 2021 Jun 3;9:e11565. doi: 10.7717/peerj.11565. eCollection 2021. PeerJ. 2021. PMID: 34141493 Free PMC article.
-
Mixed Infections and Rifampin Heteroresistance among Mycobacterium tuberculosis Clinical Isolates.J Clin Microbiol. 2015 Jul;53(7):2138-47. doi: 10.1128/JCM.03507-14. Epub 2015 Apr 22. J Clin Microbiol. 2015. PMID: 25903578 Free PMC article.
-
Sub-Lineage Specific Phenolic Glycolipid Patterns in the Mycobacterium tuberculosis Complex Lineage 1.Front Microbiol. 2022 Mar 8;13:832054. doi: 10.3389/fmicb.2022.832054. eCollection 2022. Front Microbiol. 2022. PMID: 35350619 Free PMC article.
-
In vivo virulence of Mycobacterium tuberculosis depends on a single homologue of the LytR-CpsA-Psr proteins.Sci Rep. 2018 Mar 2;8(1):3936. doi: 10.1038/s41598-018-22012-6. Sci Rep. 2018. PMID: 29500450 Free PMC article.
References
-
- WHO 2012. Global tuberculosis report 2012. WHO, Geneva, Switzerland
-
- Gagneux S, Small PM. 2007. Global phylogeography of Mycobacterium tuberculosis and implications for tuberculosis product development. Lancet Infect. Dis. 7:328–337 - PubMed
-
- Wirth T, Hildebrand F, Allix-Béguec C, Wölbeling F, Kubica T, Kremer K, van Soolingen D, Rüsch-Gerdes S, Locht C, Brisse S, Meyer A, Supply P, Niemann S. 2008. Origin, spread and demography of the Mycobacterium tuberculosis complex. PLoS Pathog. 4:e1000160 http://dx.doi.org/doi:10.1371/journal.ppat.1000160 - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
