Barrier to autointegration factor (BAF) inhibits vaccinia virus intermediate transcription in the absence of the viral B1 kinase
- PMID: 23891157
- PMCID: PMC3755115
- DOI: 10.1016/j.virol.2013.07.002
Barrier to autointegration factor (BAF) inhibits vaccinia virus intermediate transcription in the absence of the viral B1 kinase
Abstract
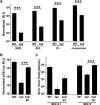
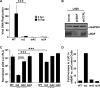
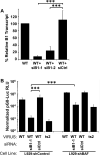
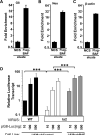
Barrier to autointegration factor (BAF/BANF1) is a cellular DNA-binding protein found in the nucleus and cytoplasm. Cytoplasmic BAF binds to foreign DNA and can act as a defense against vaccinia DNA replication. To evade BAF, vaccinia expresses the B1 kinase, which phosphorylates BAF and blocks its ability to bind DNA. Interestingly, B1 is also needed for viral intermediate gene expression via an unknown mechanism. Therefore, we evaluated the impact of B1-BAF signaling on vaccinia transcription. Strikingly, the decrease in vaccinia transcription caused by loss of B1 can be rescued by depletion of BAF. The repressive action of BAF is greatest on a viral promoter, and is more modest when non-vaccinia promoters are employed, which suggests BAF acts in a gene specific manner. These studies expand our understanding of the role of the B1 kinase during infection and provide the first evidence that BAF is a defense against viral gene expression.
Keywords: BAF; BANF1; Barrier to autointegration; Transcription; Vaccinia; Virus host interaction.
© 2013 Elsevier Inc. All rights reserved.
Figures
Similar articles
-
Diverse cellular functions of barrier-to-autointegration factor and its roles in disease.J Cell Sci. 2020 Aug 17;133(16):jcs246546. doi: 10.1242/jcs.246546. J Cell Sci. 2020. PMID: 32817163 Free PMC article. Review.
-
The Vaccinia Virus B12 Pseudokinase Represses Viral Replication via Interaction with the Cellular Kinase VRK1 and Activation of the Antiviral Effector BAF.J Virol. 2021 Jan 13;95(3):e02114-20. doi: 10.1128/JVI.02114-20. Print 2021 Jan 13. J Virol. 2021. PMID: 33177193 Free PMC article.
-
Poxviral B1 kinase overcomes barrier to autointegration factor, a host defense against virus replication.Cell Host Microbe. 2007 May 17;1(3):187-97. doi: 10.1016/j.chom.2007.03.007. Cell Host Microbe. 2007. PMID: 18005698 Free PMC article.
-
Vaccinia Virus B1 Kinase Is Required for Postreplicative Stages of the Viral Life Cycle in a BAF-Independent Manner in U2OS Cells.J Virol. 2015 Oct;89(20):10247-59. doi: 10.1128/JVI.01252-15. Epub 2015 Jul 29. J Virol. 2015. PMID: 26223647 Free PMC article.
-
Barrier-to-autointegration factor--a BAFfling little protein.Trends Cell Biol. 2007 Apr;17(4):202-8. doi: 10.1016/j.tcb.2007.02.004. Epub 2007 Feb 21. Trends Cell Biol. 2007. PMID: 17320395 Review.
Cited by
-
Diverse cellular functions of barrier-to-autointegration factor and its roles in disease.J Cell Sci. 2020 Aug 17;133(16):jcs246546. doi: 10.1242/jcs.246546. J Cell Sci. 2020. PMID: 32817163 Free PMC article. Review.
-
Abortive Infection of Animal Cells: What Goes Wrong.Annu Rev Virol. 2024 Sep;11(1):193-213. doi: 10.1146/annurev-virology-100422-023037. Epub 2024 Aug 30. Annu Rev Virol. 2024. PMID: 38631917 Review.
-
The Vaccinia Virus B12 Pseudokinase Represses Viral Replication via Interaction with the Cellular Kinase VRK1 and Activation of the Antiviral Effector BAF.J Virol. 2021 Jan 13;95(3):e02114-20. doi: 10.1128/JVI.02114-20. Print 2021 Jan 13. J Virol. 2021. PMID: 33177193 Free PMC article.
-
Deletion of the Vaccinia Virus B1 Kinase Reveals Essential Functions of This Enzyme Complemented Partly by the Homologous Cellular Kinase VRK2.J Virol. 2017 Jul 12;91(15):e00635-17. doi: 10.1128/JVI.00635-17. Print 2017 Aug 1. J Virol. 2017. PMID: 28515294 Free PMC article.
-
The TFAMoplex-Conversion of the Mitochondrial Transcription Factor A into a DNA Transfection Agent.Adv Sci (Weinh). 2022 Mar;9(8):e2104987. doi: 10.1002/advs.202104987. Epub 2022 Jan 17. Adv Sci (Weinh). 2022. PMID: 35038234 Free PMC article.
References
-
- Amegadzie BY, Ahn BY, Moss B. Identification, sequence, and expression of the gene encoding a Mr 35,000 subunit of the vaccinia virus DNA-dependent RNA polymerase. Journal of Biological Chemistry. 1991;266:13712–13718. - PubMed
-
- Banham AH, Smith GL. Vaccinia virus gene B1R encodes a 34-kDa serine/threonine protein kinase that localizes in cytoplasmic factories and is packaged into virions. Virology. 1992;191:803–812. - PubMed
-
- Black EP, Moussatche N, Condit RC. Characterization of the interactions among vaccinia virus transcription factors G2R, A18R, and H5R. Virology. 1998;245:313–322. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

