Preparation of biologically active Arabidopsis ribosomes and comparison with yeast ribosomes for binding to a tRNA-mimic that enhances translation of plant plus-strand RNA viruses
- PMID: 23885260
- PMCID: PMC3718319
- DOI: 10.3389/fpls.2013.00271
Preparation of biologically active Arabidopsis ribosomes and comparison with yeast ribosomes for binding to a tRNA-mimic that enhances translation of plant plus-strand RNA viruses
Abstract
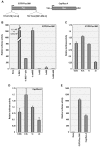
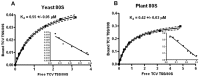
Isolation of biologically active cell components from multicellular eukaryotic organisms often poses difficult challenges such as low yields and inability to retain the integrity and functionality of the purified compound. We previously identified a cap-independent translation enhancer (3'CITE) in the 3'UTR of Turnip crinkle virus (TCV) that structurally mimics a tRNA and binds to yeast 80S ribosomes and 60S subunits in the P-site. Yeast ribosomes were used for these studies due to the lack of methods for isolation of plant ribosomes with high yields and integrity. To carry out studies with more natural components, a simple and efficient procedure has been developed for the isolation of large quantities of high quality ribosomes and ribosomal subunits from Arabidopsis thaliana protoplasts prepared from seed-derived callus tissue. Attempts to isolate high quality ribosomes from wheat germ, bean sprouts, and evacuolated protoplasts were unsuccessful. Addition of purified Arabidopsis 80S plant ribosomes to ribosome-depleted wheat germ lysates resulted in a greater than 1200-fold enhancement in in vitro translation of a luciferase reporter construct. The TCV 3'CITE bound to ribosomes with a three to sevenfold higher efficiency when using plant 80S ribosomes compared with yeast ribosomes, indicating that this viral translational enhancer is adapted to interact more efficiently with host plant ribosomes.
Keywords: 3′CITE; Arabidopsis thaliana protoplasts; TCV; TSS; plant ribosomes; virus translation.
Figures


Similar articles
-
The kissing-loop T-shaped structure translational enhancer of Pea enation mosaic virus can bind simultaneously to ribosomes and a 5' proximal hairpin.J Virol. 2013 Nov;87(22):11987-2002. doi: 10.1128/JVI.02005-13. Epub 2013 Aug 28. J Virol. 2013. PMID: 23986599 Free PMC article.
-
The 3' untranslated region of Pea Enation Mosaic Virus contains two T-shaped, ribosome-binding, cap-independent translation enhancers.J Virol. 2014 Oct;88(20):11696-712. doi: 10.1128/JVI.01433-14. Epub 2014 Aug 6. J Virol. 2014. PMID: 25100834 Free PMC article.
-
The 3' proximal translational enhancer of Turnip crinkle virus binds to 60S ribosomal subunits.RNA. 2008 Nov;14(11):2379-93. doi: 10.1261/rna.1227808. Epub 2008 Sep 29. RNA. 2008. PMID: 18824512 Free PMC article.
-
3'UTRs of carmoviruses.Virus Res. 2015 Aug 3;206:27-36. doi: 10.1016/j.virusres.2015.01.023. Epub 2015 Feb 4. Virus Res. 2015. PMID: 25662021 Review.
-
3' Cap-independent translation enhancers of positive-strand RNA plant viruses.Curr Opin Virol. 2011 Nov;1(5):373-80. doi: 10.1016/j.coviro.2011.10.002. Epub 2011 Oct 24. Curr Opin Virol. 2011. PMID: 22440838 Review.
Cited by
-
The kissing-loop T-shaped structure translational enhancer of Pea enation mosaic virus can bind simultaneously to ribosomes and a 5' proximal hairpin.J Virol. 2013 Nov;87(22):11987-2002. doi: 10.1128/JVI.02005-13. Epub 2013 Aug 28. J Virol. 2013. PMID: 23986599 Free PMC article.
-
The 3' untranslated region of Pea Enation Mosaic Virus contains two T-shaped, ribosome-binding, cap-independent translation enhancers.J Virol. 2014 Oct;88(20):11696-712. doi: 10.1128/JVI.01433-14. Epub 2014 Aug 6. J Virol. 2014. PMID: 25100834 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

