Small ruminant lentiviruses (SRLVs) break the species barrier to acquire new host range
- PMID: 23881276
- PMCID: PMC3738966
- DOI: 10.3390/v5071867
Small ruminant lentiviruses (SRLVs) break the species barrier to acquire new host range
Abstract
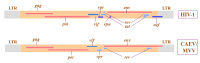
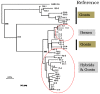
Zoonotic events of simian immunodeficiency virus (SIV) from non-human primates to humans have generated the acquired immunodeficiency syndrome (AIDS), one of the most devastating infectious disease of the last century with more than 30 million people dead and about 40.3 million people currently infected worldwide. Human immunodeficiency virus (HIV-1 and HIV-2), the two major viruses that cause AIDS in humans are retroviruses of the lentivirus genus. The genus includes arthritis-encephalitis virus (CAEV) and Maedi-Visna virus (MVV), and a heterogeneous group of viruses known as small ruminant lentiviruses (SRLVs), affecting goat and sheep. Lentivirus genome integrates into the host DNA, causing persistent infection associated with a remarkable diversity during viral replication. Direct evidence of mixed infections with these two closely related SRLVs was found in both sheep and goats. The evidence of a genetic continuum with caprine and ovine field isolates demonstrates the absence of an efficient species barrier preventing cross-species transmission. In dual-infected animals, persistent infections with both CAEV and MVV have been described, and viral chimeras have been detected. This not only complicates animal trade between countries but favors the risk that highly pathogenic variants may emerge as has already been observed in the past in Iceland and, more recently, in outbreaks with virulent strains in Spain. SRLVs affecting wildlife have already been identified, demonstrating the existence of emergent viruses adapted to new hosts. Viruses adapted to wildlife ruminants may acquire novel biopathological properties which may endanger not only the new host species but also domestic ruminants and humans. SRLVs infecting sheep and goats follow a genomic evolution similar to that observed in HIV or in other lentiviruses. Lentivirus genetic diversity and host factors leading to the establishment of naturally occurring virulent versus avirulent infections, in addition to the emergence of new strains, challenge every aspect of SRLV control measures for providing efficient tools to prevent the transmission of diseases between wild ungulates and livestock.
Figures
Similar articles
-
Genetic characterization of small ruminant lentiviruses circulating in naturally infected sheep and goats in Ontario, Canada.Virus Res. 2013 Jul;175(1):30-44. doi: 10.1016/j.virusres.2013.03.019. Epub 2013 Apr 10. Virus Res. 2013. PMID: 23583225
-
Demonstration of coinfection with and recombination by caprine arthritis-encephalitis virus and maedi-visna virus in naturally infected goats.J Virol. 2007 May;81(10):4948-55. doi: 10.1128/JVI.00126-07. Epub 2007 Mar 7. J Virol. 2007. PMID: 17344293 Free PMC article.
-
Virological and phylogenetic characterization of attenuated small ruminant lentivirus isolates eluding efficient serological detection.Vet Microbiol. 2013 Mar 23;162(2-4):572-581. doi: 10.1016/j.vetmic.2012.11.017. Epub 2012 Nov 14. Vet Microbiol. 2013. PMID: 23206411
-
SRLVs: a genetic continuum of lentiviral species in sheep and goats with cumulative evidence of cross species transmission.Curr HIV Res. 2010 Jan;8(1):94-100. doi: 10.2174/157016210790416415. Curr HIV Res. 2010. PMID: 20210785 Review.
-
Immunogenetics of small ruminant lentiviral infections.Viruses. 2014 Aug 22;6(8):3311-33. doi: 10.3390/v6083311. Viruses. 2014. PMID: 25153344 Free PMC article. Review.
Cited by
-
Small ruminant lentivirus infection influences expression of acute phase proteins and cathelicidin genes in milk somatic cells and peripheral blood leukocytes of dairy goats.Vet Res. 2018 Nov 13;49(1):113. doi: 10.1186/s13567-018-0607-x. Vet Res. 2018. PMID: 30424807 Free PMC article.
-
Achievements of an eradication programme against caprine arthritis encephalitis virus in South Tyrol, Italy.Vet Rec. 2018 Jan 13;182(2):51. doi: 10.1136/vr.104503. Epub 2017 Nov 6. Vet Rec. 2018. PMID: 29109181 Free PMC article.
-
Serological Survey of Small Ruminant Lentivirus Infections in Free-Ranging Mouflon and Chamois in Slovenia.Animals (Basel). 2022 Apr 15;12(8):1032. doi: 10.3390/ani12081032. Animals (Basel). 2022. PMID: 35454279 Free PMC article.
-
Lentivirus Susceptibility in Brazilian and US Sheep with TMEM154 Mutations.Genes (Basel). 2022 Dec 26;14(1):70. doi: 10.3390/genes14010070. Genes (Basel). 2022. PMID: 36672811 Free PMC article.
-
First survey on association of TMEM154 and CCR5 variants with serological maedi-visna status of sheep in German flocks.Vet Res. 2018 Apr 19;49(1):36. doi: 10.1186/s13567-018-0533-y. Vet Res. 2018. PMID: 29673399 Free PMC article.
References
-
- Thorma H. Maedi-visna virus and its relationship to human immunodeficiency virus. AIDS Rev. 2005;7:233–245. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

