Male-biased genes in catfish as revealed by RNA-Seq analysis of the testis transcriptome
- PMID: 23874634
- PMCID: PMC3709890
- DOI: 10.1371/journal.pone.0068452
Male-biased genes in catfish as revealed by RNA-Seq analysis of the testis transcriptome
Abstract
Background: Catfish has a male-heterogametic (XY) sex determination system, but genes involved in gonadogenesis, spermatogenesis, testicular determination, and sex determination are poorly understood. As a first step of understanding the transcriptome of the testis, here, we conducted RNA-Seq analysis using high throughput Illumina sequencing.
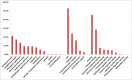
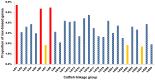
Methodology/principal findings: A total of 269.6 million high quality reads were assembled into 193,462 contigs with a N50 length of 806 bp. Of these contigs, 67,923 contigs had hits to a set of 25,307 unigenes, including 167 unique genes that had not been previously identified in catfish. A meta-analysis of expressed genes in the testis and in the gynogen (double haploid female) allowed the identification of 5,450 genes that are preferentially expressed in the testis, providing a pool of putative male-biased genes. Gene ontology and annotation analysis suggested that many of these male-biased genes were involved in gonadogenesis, spermatogenesis, testicular determination, gametogenesis, gonad differentiation, and possibly sex determination.
Conclusion/significance: We provide the first transcriptome-level analysis of the catfish testis. Our analysis would lay the basis for sequential follow-up studies of genes involved in sex determination and differentiation in catfish.
Conflict of interest statement
Figures
Similar articles
-
Gonadal transcriptomic analysis of yellow catfish (Pelteobagrus fulvidraco): identification of sex-related genes and genetic markers.Physiol Genomics. 2014 Nov 1;46(21):798-807. doi: 10.1152/physiolgenomics.00088.2014. Epub 2014 Sep 2. Physiol Genomics. 2014. PMID: 25185028
-
De novo transcriptome assembly and sex-biased gene expression in the gonads of Amur catfish (Silurus asotus).Genomics. 2020 May;112(3):2603-2614. doi: 10.1016/j.ygeno.2020.01.026. Epub 2020 Feb 25. Genomics. 2020. PMID: 32109564
-
Efficient assembly and annotation of the transcriptome of catfish by RNA-Seq analysis of a doubled haploid homozygote.BMC Genomics. 2012 Nov 5;13:595. doi: 10.1186/1471-2164-13-595. BMC Genomics. 2012. PMID: 23127152 Free PMC article.
-
A comprehensive transcriptome provides candidate genes for sex determination/differentiation and SSR/SNP markers in yellow catfish.Mar Biotechnol (NY). 2015 Apr;17(2):190-8. doi: 10.1007/s10126-014-9607-7. Epub 2014 Nov 18. Mar Biotechnol (NY). 2015. PMID: 25403497
-
Transcriptome analysis of starch and sucrose metabolism across bulb development in Sagittaria sagittifolia.Gene. 2018 Apr 5;649:99-112. doi: 10.1016/j.gene.2018.01.075. Epub 2018 Jan 31. Gene. 2018. PMID: 29374598 Review.
Cited by
-
Advances in Reproductive Endocrinology and Neuroendocrine Research Using Catfish Models.Cells. 2021 Oct 20;10(11):2807. doi: 10.3390/cells10112807. Cells. 2021. PMID: 34831032 Free PMC article. Review.
-
The Gene Toolkit Implicated in Functional Sex in Sparidae Hermaphrodites: Inferences From Comparative Transcriptomics.Front Genet. 2019 Jan 18;9:749. doi: 10.3389/fgene.2018.00749. eCollection 2018. Front Genet. 2019. PMID: 30713551 Free PMC article.
-
Transcriptome Analysis of Male and Female Mature Gonads of Silver Sillago (Sillago sihama).Genes (Basel). 2019 Feb 11;10(2):129. doi: 10.3390/genes10020129. Genes (Basel). 2019. PMID: 30754713 Free PMC article.
-
Comparison of Gonadal Transcriptomes Uncovers Reproduction-Related Genes with Sexually Dimorphic Expression Patterns in Diodon hystrix.Animals (Basel). 2021 Apr 7;11(4):1042. doi: 10.3390/ani11041042. Animals (Basel). 2021. PMID: 33917262 Free PMC article.
-
Identification of an Epigenetically Marked Locus within the Sex Determination Region of Channel Catfish.Int J Mol Sci. 2022 May 13;23(10):5471. doi: 10.3390/ijms23105471. Int J Mol Sci. 2022. PMID: 35628283 Free PMC article.
References
-
- de Mitcheson YS, Liu M (2008) Functional hermaphroditism in teleosts. Fish and Fisheries 9: 1–43.
-
- Kobayashi Y, Nagahama Y, Nakamura M (2013) Diversity and plasticity of sex determination and differentiation in fishes. Sex Dev 7: 115–125. - PubMed
-
- Devlin RH, Nagahama Y (2002) Sex determination and sex differentiation in fish: an overview of genetic, physiological, and environmental influences. Aquaculture 208: 191–364.
-
- Matsuda M, Nagahama Y, Shinomiya A, Sato T, Matsuda C, et al. (2002) DMY is a Y-specific DM-domain gene required for male development in the medaka fish. Nature 417: 559–563. - PubMed
-
- Yano A, Guyomard R, Nicol B, Jouanno E, Quillet E, et al. (2012) An Immune-Related Gene Evolved into the Master Sex-Determining Gene in Rainbow Trout, Oncorhynchus mykiss. Current Biology 22: 1423–1428. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

