Genomic analysis of natural selection and phenotypic variation in high-altitude mongolians
- PMID: 23874230
- PMCID: PMC3715426
- DOI: 10.1371/journal.pgen.1003634
Genomic analysis of natural selection and phenotypic variation in high-altitude mongolians
Abstract
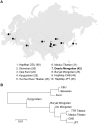
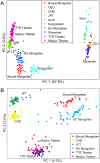
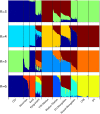
Deedu (DU) Mongolians, who migrated from the Mongolian steppes to the Qinghai-Tibetan Plateau approximately 500 years ago, are challenged by environmental conditions similar to native Tibetan highlanders. Identification of adaptive genetic factors in this population could provide insight into coordinated physiological responses to this environment. Here we examine genomic and phenotypic variation in this unique population and present the first complete analysis of a Mongolian whole-genome sequence. High-density SNP array data demonstrate that DU Mongolians share genetic ancestry with other Mongolian as well as Tibetan populations, specifically in genomic regions related with adaptation to high altitude. Several selection candidate genes identified in DU Mongolians are shared with other Asian groups (e.g., EDAR), neighboring Tibetan populations (including high-altitude candidates EPAS1, PKLR, and CYP2E1), as well as genes previously hypothesized to be associated with metabolic adaptation (e.g., PPARG). Hemoglobin concentration, a trait associated with high-altitude adaptation in Tibetans, is at an intermediate level in DU Mongolians compared to Tibetans and Han Chinese at comparable altitude. Whole-genome sequence from a DU Mongolian (Tianjiao1) shows that about 2% of the genomic variants, including more than 300 protein-coding changes, are specific to this individual. Our analyses of DU Mongolians and the first Mongolian genome provide valuable insight into genetic adaptation to extreme environments.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
Similar articles
-
Identification of a Tibetan-specific mutation in the hypoxic gene EGLN1 and its contribution to high-altitude adaptation.Mol Biol Evol. 2013 Aug;30(8):1889-98. doi: 10.1093/molbev/mst090. Epub 2013 May 10. Mol Biol Evol. 2013. PMID: 23666208
-
Differentiated demographic histories and local adaptations between Sherpas and Tibetans.Genome Biol. 2017 Jun 15;18(1):115. doi: 10.1186/s13059-017-1242-y. Genome Biol. 2017. PMID: 28619099 Free PMC article.
-
Genetic variations in Tibetan populations and high-altitude adaptation at the Himalayas.Mol Biol Evol. 2011 Feb;28(2):1075-81. doi: 10.1093/molbev/msq290. Epub 2010 Oct 28. Mol Biol Evol. 2011. PMID: 21030426
-
Adaptive genetic changes related to haemoglobin concentration in native high-altitude Tibetans.Exp Physiol. 2015 Nov;100(11):1263-8. doi: 10.1113/EP085035. Exp Physiol. 2015. PMID: 26454145 Review.
-
The Qinghai-Tibetan plateau: how high do Tibetans live?High Alt Med Biol. 2001 Winter;2(4):489-99. doi: 10.1089/152702901753397054. High Alt Med Biol. 2001. PMID: 11809089 Review.
Cited by
-
High-Altitude Adaptation: Mechanistic Insights from Integrated Genomics and Physiology.Mol Biol Evol. 2021 Jun 25;38(7):2677-2691. doi: 10.1093/molbev/msab064. Mol Biol Evol. 2021. PMID: 33751123 Free PMC article. Review.
-
Population History and Altitude-Related Adaptation in the Sherpa.Front Physiol. 2019 Aug 28;10:1116. doi: 10.3389/fphys.2019.01116. eCollection 2019. Front Physiol. 2019. PMID: 31555147 Free PMC article. Review.
-
Dissecting dynamics and differences of selective pressures in the evolution of human pigmentation.Biol Open. 2021 Feb 9;10(2):bio056523. doi: 10.1242/bio.056523. Biol Open. 2021. PMID: 33495209 Free PMC article.
-
Complex trait susceptibilities and population diversity in a sample of 4,145 Russians.Nat Commun. 2024 Jul 23;15(1):6212. doi: 10.1038/s41467-024-50304-1. Nat Commun. 2024. PMID: 39043636 Free PMC article.
-
Genetic structure in the Sherpa and neighboring Nepalese populations.BMC Genomics. 2017 Jan 19;18(1):102. doi: 10.1186/s12864-016-3469-5. BMC Genomics. 2017. PMID: 28103797 Free PMC article.
References
-
- Keyser-Tracqui C, Crubezy E, Pamzsav H, Varga T, Ludes B (2006) Population origins in Mongolia: genetic structure analysis of ancient and modern DNA. Am J Phys Anthropol 131: 272–281. - PubMed
-
- Nasidze I, Quinque D, Dupanloup I, Cordaux R, Kokshunova L, et al. (2005) Genetic evidence for the Mongolian ancestry of Kalmyks. Am J Phys Anthropol 128: 846–854. - PubMed
-
- Hei L, Tie Z (2011) Shine Nairuulsan Mongol Undusten-ne dobch tuuh (Mongolian History): Inner Mongolian People Publishing Agency.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

