Evolution of plant HECT ubiquitin ligases
- PMID: 23869223
- PMCID: PMC3712016
- DOI: 10.1371/journal.pone.0068536
Evolution of plant HECT ubiquitin ligases
Abstract
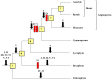
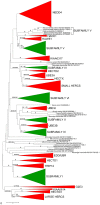
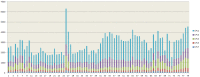
HECT ubiquitin ligases are key components of the ubiquitin-proteasome system, which is present in all eukaryotes. In this study, the patterns of emergence of HECT genes in plants are described. Phylogenetic and structural data indicate that viridiplantae have six main HECT subfamilies, which arose before the split that separated green algae from the rest of plants. It is estimated that the common ancestor of all plants contained seven HECT genes. Contrary to what happened in animals, the number of HECT genes has been kept quite constant in all lineages, both in chlorophyta and streptophyta, although evolutionary recent duplications are found in some species. Several of the genes found in plants may have originated very early in eukaryotic evolution, given that they have clear similarities, both in sequence and structure, to animal genes. Finally, in Arabidopsis thaliana, we found significant correlations in the expression patterns of HECT genes and some ancient, broadly expressed genes that belong to a different ubiquitin ligase family, called RBR. These results are discussed in the context of the evolution of the gene families required for ubiquitination in plants.
Conflict of interest statement
Figures






Similar articles
-
Diversification and Specialization of Plant RBR Ubiquitin Ligases.PLoS One. 2010 Jul 14;5(7):e11579. doi: 10.1371/journal.pone.0011579. PLoS One. 2010. PMID: 20644651 Free PMC article.
-
Origin and evolution of fungal HECT ubiquitin ligases.Sci Rep. 2018 Apr 23;8(1):6419. doi: 10.1038/s41598-018-24914-x. Sci Rep. 2018. PMID: 29686411 Free PMC article.
-
A genomic survey of HECT ubiquitin ligases in eukaryotes reveals independent expansions of the HECT system in several lineages.Genome Biol Evol. 2013;5(5):833-47. doi: 10.1093/gbe/evt052. Genome Biol Evol. 2013. PMID: 23563970 Free PMC article.
-
ATLs and BTLs, plant-specific and general eukaryotic structurally-related E3 ubiquitin ligases.Plant Sci. 2014 Feb;215-216:69-75. doi: 10.1016/j.plantsci.2013.10.017. Epub 2013 Nov 5. Plant Sci. 2014. PMID: 24388516 Review.
-
Tracking of Ubiquitin Signaling through 3.5 Billion Years of Combinatorial Conjugation.Int J Mol Sci. 2024 Aug 8;25(16):8671. doi: 10.3390/ijms25168671. Int J Mol Sci. 2024. PMID: 39201358 Free PMC article. Review.
Cited by
-
Genome-wide identification, phylogenetic and expression analysis of the maize HECT E3 ubiquitin ligase genes.Genetica. 2019 Dec;147(5-6):391-400. doi: 10.1007/s10709-019-00080-4. Epub 2019 Nov 18. Genetica. 2019. PMID: 31741104
-
An ORFeome of rice E3 ubiquitin ligases for global analysis of the ubiquitination interactome.Genome Biol. 2022 Jul 11;23(1):154. doi: 10.1186/s13059-022-02717-8. Genome Biol. 2022. PMID: 35821048 Free PMC article.
-
Suppression of Arabidopsis AtPUB30 resulted in increased tolerance to salt stress during germination.Plant Cell Rep. 2015 Feb;34(2):277-89. doi: 10.1007/s00299-014-1706-4. Epub 2014 Nov 20. Plant Cell Rep. 2015. PMID: 25410251
-
OsPUB41, a U-box E3 ubiquitin ligase, acts as a negative regulator of drought stress response in rice (Oryza Sativa L.).Plant Mol Biol. 2021 Jul;106(4-5):463-477. doi: 10.1007/s11103-021-01158-4. Epub 2021 Jun 7. Plant Mol Biol. 2021. PMID: 34100185
-
Genome-Wide Identification, Characterization, and Transcriptional Profile of the HECT E3 Ubiquitin Ligase Gene Family in the Hard-Shelled Mussel Mytilus coruscus Gould.Genes (Basel). 2024 Aug 16;15(8):1085. doi: 10.3390/genes15081085. Genes (Basel). 2024. PMID: 39202444 Free PMC article.
References
-
- Glickman MH, Ciechanover A (2002) The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol Rev 82: 373–428. - PubMed
-
- Kerscher O, Felberbaum R, Hochstrasser M (2006) Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu Rev Cell Dev Biol 22: 159–180. - PubMed
-
- Chen ZJ, Sun LJ (2009) Nonproteolytic functions of ubiquitin in cell signaling. Mol Cell 33: 275–286. - PubMed
-
- Komander D (2009) The emerging complexity of protein ubiquitination. Biochem Soc Trans 37: 937–953. - PubMed
-
- Schwartz AL, Ciechanover A (2009) Targeting proteins for destruction by the ubiquitin system: implications for human pathobiology. Annu Rev Pharmacol Toxicol 49: 73–96. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

