MREdictor: a two-step dynamic interaction model that accounts for mRNA accessibility and Pumilio binding accurately predicts microRNA targets
- PMID: 23863844
- PMCID: PMC3794589
- DOI: 10.1093/nar/gkt629
MREdictor: a two-step dynamic interaction model that accounts for mRNA accessibility and Pumilio binding accurately predicts microRNA targets
Abstract
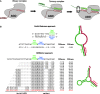
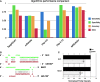
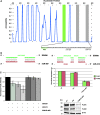
The prediction of pairing between microRNAs (miRNAs) and the miRNA recognition elements (MREs) on mRNAs is expected to be an important tool for understanding gene regulation. Here, we show that mRNAs that contain Pumilio recognition elements (PRE) in the proximity of predicted miRNA-binding sites are more likely to form stable secondary structures within their 3'-UTR, and we demonstrated using a PUM1 and PUM2 double knockdown that Pumilio proteins are general regulators of miRNA accessibility. On the basis of these findings, we developed a computational method for predicting miRNA targets that accounts for the presence of PRE in the proximity of seed-match sequences within poorly accessible structures. Moreover, we implement the miRNA-MRE duplex pairing as a two-step model, which better fits the available structural data. This algorithm, called MREdictor, allows for the identification of miRNA targets in poorly accessible regions and is not restricted to a perfect seed-match; these features are not present in other computational prediction methods.
Figures



Similar articles
-
Analysis of microRNA-target interactions by a target structure based hybridization model.Pac Symp Biocomput. 2008:64-74. Pac Symp Biocomput. 2008. PMID: 18232104
-
Efficient 3'-pairing renders microRNA targeting less sensitive to mRNA seed accessibility.Nucleic Acids Res. 2023 Nov 10;51(20):11162-11177. doi: 10.1093/nar/gkad795. Nucleic Acids Res. 2023. PMID: 37819016 Free PMC article.
-
Analysis of the accessibility of CLIP bound sites reveals that nucleation of the miRNA:mRNA pairing occurs preferentially at the 3'-end of the seed match.RNA. 2012 Oct;18(10):1760-70. doi: 10.1261/rna.033282.112. Epub 2012 Aug 22. RNA. 2012. PMID: 22915600 Free PMC article.
-
Desperately seeking microRNA targets.Nat Struct Mol Biol. 2010 Oct;17(10):1169-74. doi: 10.1038/nsmb.1921. Nat Struct Mol Biol. 2010. PMID: 20924405 Review.
-
Experimental strategies for microRNA target identification.Nucleic Acids Res. 2011 Sep 1;39(16):6845-53. doi: 10.1093/nar/gkr330. Epub 2011 Jun 7. Nucleic Acids Res. 2011. PMID: 21652644 Free PMC article. Review.
Cited by
-
Small RNA Targets: Advances in Prediction Tools and High-Throughput Profiling.Biology (Basel). 2022 Dec 11;11(12):1798. doi: 10.3390/biology11121798. Biology (Basel). 2022. PMID: 36552307 Free PMC article. Review.
-
Pumilio directs deadenylation-associated translational repression of the cyclin-dependent kinase 1 activator RGC-32.Nucleic Acids Res. 2018 Apr 20;46(7):3707-3725. doi: 10.1093/nar/gky038. Nucleic Acids Res. 2018. PMID: 29385536 Free PMC article.
-
Scan for Motifs: a webserver for the analysis of post-transcriptional regulatory elements in the 3' untranslated regions (3' UTRs) of mRNAs.BMC Bioinformatics. 2014 Jun 8;15:174. doi: 10.1186/1471-2105-15-174. BMC Bioinformatics. 2014. PMID: 24909639 Free PMC article.
-
MicroRNAs-143 and -145 induce epithelial to mesenchymal transition and modulate the expression of junction proteins.Cell Death Differ. 2017 Oct;24(10):1750-1760. doi: 10.1038/cdd.2017.103. Epub 2017 Jun 23. Cell Death Differ. 2017. PMID: 28644441 Free PMC article.
-
TET1 is a tumour suppressor that inhibits colon cancer growth by derepressing inhibitors of the WNT pathway.Oncogene. 2015 Aug 6;34(32):4168-76. doi: 10.1038/onc.2014.356. Epub 2014 Nov 3. Oncogene. 2015. PMID: 25362856
References
-
- Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. - PubMed
-
- Frank F, Sonenberg N, Nagar B. Structural basis for 5′-nucleotide base-specific recognition of guide RNA by human AGO2. Nature. 2010;465:818–822. - PubMed
-
- Brodersen P, Voinnet O. Revisiting the principles of microRNA target recognition and mode of action. Nat. Rev. Mol. Cell Biol. 2009;10:141–148. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

