Physiological adaptations of Saccharomyces cerevisiae evolved for improved butanol tolerance
- PMID: 23855998
- PMCID: PMC3729582
- DOI: 10.1186/1754-6834-6-101
Physiological adaptations of Saccharomyces cerevisiae evolved for improved butanol tolerance
Abstract
Background: Butanol is a chemical with potential uses as biofuel and solvent, which can be produced by microbial fermentation. However, the end product toxicity is one of the main obstacles for developing the production process irrespective of the choice of production organism. The long-term goal of the present project is to produce 2-butanol in Saccharomyces cerevisiae. Therefore, unraveling the toxicity mechanisms of solvents such as butanol and understanding the mechanisms by which tolerant strains of S. cerevisiae adapt to them would be an important contribution to the development of a bio-based butanol production process.
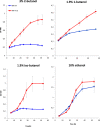
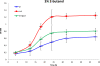
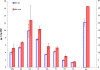
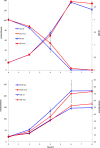
Results: A butanol tolerant S. cerevisiae was achieved through a series of sequential batch cultures with gradual increase of 2-butanol concentration. The final mutant (JBA-mut) tolerates all different alcohols tested at higher concentrations compared to the wild type (JBA-wt). Proteomics analysis of the two strains grown under mild butanol-stress revealed 46 proteins changing their expression by more than 1.5-fold in JBA-mut, 34 of which were upregulated. Strikingly, 21 out of the 34 upregulated proteins were predicted constituents of mitochondria. Among the non-mitochondrial up-regulated proteins, the minor isoform of Glycerol-3-phosphatase (Gpp2) was the most notable, since it was the only tested protein whose overexpression was found to confer butanol tolerance.
Conclusion: The study demonstrates several differences between the butanol tolerant mutant and the wild type. Upregulation of proteins involved in the mitochondrial ATP synthesizing machinery constituents and glycerol biosynthesis seem to be beneficial for a successful adaptation of yeast cells to butanol stress.
Figures
Similar articles
-
Phenotypic characterisation of Saccharomyces spp. for tolerance to 1-butanol.J Ind Microbiol Biotechnol. 2014 Nov;41(11):1627-36. doi: 10.1007/s10295-014-1511-7. Epub 2014 Sep 23. J Ind Microbiol Biotechnol. 2014. PMID: 25242291
-
Genome-scale analyses of butanol tolerance in Saccharomyces cerevisiae reveal an essential role of protein degradation.Biotechnol Biofuels. 2013 Apr 3;6(1):48. doi: 10.1186/1754-6834-6-48. Biotechnol Biofuels. 2013. PMID: 23552365 Free PMC article.
-
Butanol production by Saccharomyces cerevisiae: perspectives, strategies and challenges.World J Microbiol Biotechnol. 2020 Mar 9;36(3):48. doi: 10.1007/s11274-020-02828-z. World J Microbiol Biotechnol. 2020. PMID: 32152786 Review.
-
Producing alcohol and salt stress tolerant strain of Saccharomyces cerevisiae by heterologous expression of pprI gene.Enzyme Microb Technol. 2019 May;124:17-22. doi: 10.1016/j.enzmictec.2019.01.008. Epub 2019 Jan 23. Enzyme Microb Technol. 2019. PMID: 30797475
-
Prospective and development of butanol as an advanced biofuel.Biotechnol Adv. 2013 Dec;31(8):1575-84. doi: 10.1016/j.biotechadv.2013.08.004. Epub 2013 Aug 27. Biotechnol Adv. 2013. PMID: 23993946 Review.
Cited by
-
Comparative sequence analysis and mutagenesis of ethylene forming enzyme (EFE) 2-oxoglutarate/Fe(II)-dependent dioxygenase homologs.BMC Biochem. 2014 Oct 2;15:22. doi: 10.1186/1471-2091-15-22. BMC Biochem. 2014. PMID: 25278273 Free PMC article.
-
Clostridium cellulovorans Proteomic Responses to Butanol Stress.Front Microbiol. 2021 Jul 21;12:674639. doi: 10.3389/fmicb.2021.674639. eCollection 2021. Front Microbiol. 2021. PMID: 34367082 Free PMC article.
-
Genotype-by-Environment-by-Environment Interactions in the Saccharomyces cerevisiae Transcriptomic Response to Alcohols and Anaerobiosis.G3 (Bethesda). 2018 Dec 10;8(12):3881-3890. doi: 10.1534/g3.118.200677. G3 (Bethesda). 2018. PMID: 30301737 Free PMC article.
-
Phenotypic characterisation of Saccharomyces spp. for tolerance to 1-butanol.J Ind Microbiol Biotechnol. 2014 Nov;41(11):1627-36. doi: 10.1007/s10295-014-1511-7. Epub 2014 Sep 23. J Ind Microbiol Biotechnol. 2014. PMID: 25242291
-
Biobutanol from cheese whey.Microb Cell Fact. 2015 Mar 5;14:27. doi: 10.1186/s12934-015-0200-1. Microb Cell Fact. 2015. PMID: 25889728 Free PMC article. Review.
References
-
- Dürre P. Biobutanol: An attractive biofuel. Biotechnol J. 2007;2:1525–1534. - PubMed
-
- Minteer S. Alcoholic fuels. 6000 Broken Sound Parkway NW, USA: CRC Press Taylor & Francis Group; 2006.
-
- Savage DF, Way J, Silver PA. Defossiling fuel: how synthetic biology can transform biofuel production. ACS Chem Biol. 2008;3:13–16. - PubMed
-
- Zverlov VV, Berezina O, Velikodvorskaya GA, Schwarz WH. Bacterial acetone and butanol production by industrial fermentation in the Soviet Union: use of hydrolyzed agricultural waste for biorefinery. Appl Microbiol Biotechnol. 2006;71:587–597. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

