GRG5/AES interacts with T-cell factor 4 (TCF4) and downregulates Wnt signaling in human cells and zebrafish embryos
- PMID: 23840876
- PMCID: PMC3698143
- DOI: 10.1371/journal.pone.0067694
GRG5/AES interacts with T-cell factor 4 (TCF4) and downregulates Wnt signaling in human cells and zebrafish embryos
Abstract
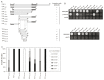
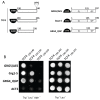
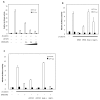
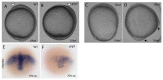
Transcriptional control by TCF/LEF proteins is crucial in key developmental processes such as embryo polarity, tissue architecture and cell fate determination. TCFs associate with β-catenin to activate transcription in the presence of Wnt signaling, but in its absence act as repressors together with Groucho-family proteins (GRGs). TCF4 is critical in vertebrate intestinal epithelium, where TCF4-β-catenin complexes are necessary for the maintenance of a proliferative compartment, and their abnormal formation initiates tumorigenesis. However, the extent of TCF4-GRG complexes' roles in development and the mechanisms by which they repress transcription are not completely understood. Here we characterize the interaction between TCF4 and GRG5/AES, a Groucho family member whose functional relationship with TCFs has been controversial. We map the core GRG interaction region in TCF4 to a 111-amino acid fragment and show that, in contrast to other GRGs, GRG5/AES-binding specifically depends on a 4-amino acid motif (LVPQ) present only in TCF3 and some TCF4 isoforms. We further demonstrate that GRG5/AES represses Wnt-mediated transcription both in human cells and zebrafish embryos. Importantly, we provide the first evidence of an inherent repressive function of GRG5/AES in dorsal-ventral patterning during early zebrafish embryogenesis. These results improve our understanding of TCF-GRG interactions, have significant implications for models of transcriptional repression by TCF-GRG complexes, and lay the groundwork for in depth direct assessment of the potential role of Groucho-family proteins in both normal and abnormal development.
Conflict of interest statement
Figures





Similar articles
-
VBP1 modulates Wnt/β-catenin signaling by mediating the stability of the transcription factors TCF/LEFs.J Biol Chem. 2020 Dec 4;295(49):16826-16839. doi: 10.1074/jbc.RA120.015282. Epub 2020 Sep 28. J Biol Chem. 2020. PMID: 32989053 Free PMC article.
-
AES/GRG5: more than just a dominant-negative TLE/GRG family member.Dev Dyn. 2010 Nov;239(11):2795-805. doi: 10.1002/dvdy.22439. Dev Dyn. 2010. PMID: 20925119 Free PMC article. Review.
-
Tissue-specific derepression of TCF/LEF controls the activity of the Wnt/β-catenin pathway.Nat Commun. 2014 Nov 5;5:5368. doi: 10.1038/ncomms6368. Nat Commun. 2014. PMID: 25371059
-
A dynamic exchange of TCF3 and TCF4 transcription factors controls MYC expression in colorectal cancer cells.Cell Cycle. 2015;14(3):323-32. doi: 10.4161/15384101.2014.980643. Cell Cycle. 2015. PMID: 25659031 Free PMC article.
-
The Yin-Yang of TCF/beta-catenin signaling.Adv Cancer Res. 2000;77:1-24. doi: 10.1016/s0065-230x(08)60783-6. Adv Cancer Res. 2000. PMID: 10549354 Review.
Cited by
-
Groucho related gene 5 (GRG5) is involved in embryonic and neural stem cell state decisions.Sci Rep. 2018 Sep 13;8(1):13790. doi: 10.1038/s41598-018-31696-9. Sci Rep. 2018. PMID: 30214018 Free PMC article.
-
Untranslated Region Sequences and the Efficacy of mRNA Vaccines against Tuberculosis.Int J Mol Sci. 2024 Jan 10;25(2):888. doi: 10.3390/ijms25020888. Int J Mol Sci. 2024. PMID: 38255961 Free PMC article.
-
Effects of Combinations of Untranslated-Region Sequences on Translation of mRNA.Biomolecules. 2023 Nov 20;13(11):1677. doi: 10.3390/biom13111677. Biomolecules. 2023. PMID: 38002359 Free PMC article.
-
Controlling the Messenger: Regulated Translation of Maternal mRNAs in Xenopus laevis Development.Adv Exp Med Biol. 2017;953:49-82. doi: 10.1007/978-3-319-46095-6_2. Adv Exp Med Biol. 2017. PMID: 27975270 Free PMC article. Review.
-
Glucocorticoid Receptor β Acts as a Co-activator of T-Cell Factor 4 and Enhances Glioma Cell Proliferation.Mol Neurobiol. 2015 Dec;52(3):1106-1118. doi: 10.1007/s12035-014-8900-9. Epub 2014 Oct 10. Mol Neurobiol. 2015. PMID: 25301232
References
-
- Castrop J, van Norren K, Clevers H (1992) A gene family of HMG-box transcription factors with homology to TCF-1. Nucleic Acids Res 20: 611. doi:10.1093/nar/20.3.611. PubMed: 1741298. - DOI - PMC - PubMed
-
- Giese K, Amsterdam A, Grosschedl R (1991) DNA-binding properties of the HMG domain of the lymphoid-specific transcriptional regulator LEF-1. Genes Dev 5: 2567-2578. doi:10.1101/gad.5.12b.2567. PubMed: 1752444. - DOI - PubMed
-
- Giese K, Cox J, Grosschedl R (1992) The HMG domain of lymphoid enhancer factor 1 bends DNA and facilitates assembly of functional nucleoprotein structures. Cell 69: 185-195. doi:10.1016/0092-8674(92)90129-Z. PubMed: 1555239. - DOI - PubMed
-
- Travis A, Amsterdam A, Belanger C, Grosschedl R (1991) LEF-1, a gene encoding a lymphoid-specific protein with an HMG domain, regulates T-cell receptor alpha enhancer function [corrected]. Genes Dev 5: 880-894. doi:10.1101/gad.5.5.880. PubMed: 1827423. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

