Comparison of lentiviral and sleeping beauty mediated αβ T cell receptor gene transfer
- PMID: 23840834
- PMCID: PMC3695921
- DOI: 10.1371/journal.pone.0068201
Comparison of lentiviral and sleeping beauty mediated αβ T cell receptor gene transfer
Abstract
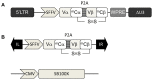
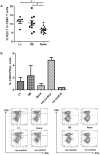
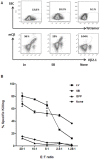
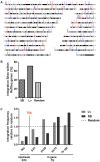
Transfer of tumour antigen-specific receptors to T cells requires efficient delivery and integration of transgenes, and currently most clinical studies are using gamma retroviral or lentiviral systems. Whilst important proof-of-principle data has been generated for both chimeric antigen receptors and αβ T cell receptors, the current platforms are costly, time-consuming and relatively inflexible. Alternative, more cost-effective, Sleeping Beauty transposon-based plasmid systems could offer a pathway to accelerated clinical testing of a more diverse repertoire of recombinant high affinity T cell receptors. Nucleofection of hyperactive SB100X transposase-mediated stable transposition of an optimised murine-human chimeric T cell receptor specific for Wilm's tumour antigen from a Sleeping Beauty transposon plasmid. Whilst transfer efficiency was lower than that mediated by lentiviral transduction, cells could be readily enriched and expanded, and mediated effective target cells lysis in vitro and in vivo. Integration sites of transposed TCR genes in primary T cells were almost randomly distributed, contrasting the predilection of lentiviral vectors for transcriptionally active sites. The results support exploitation of the Sleeping Beauty plasmid based system as a flexible and adaptable platform for accelerated, early-phase assessment of T cell receptor gene therapies.
Conflict of interest statement
Figures

Similar articles
-
A transposon and transposase system for human application.Mol Ther. 2010 Apr;18(4):674-83. doi: 10.1038/mt.2010.2. Epub 2010 Jan 26. Mol Ther. 2010. PMID: 20104209 Free PMC article. Review.
-
Stable transduction of the neonatal mouse liver using a hybrid rAAV/sleeping beauty transposon gene delivery system.J Gene Med. 2024 Aug;26(8):e3726. doi: 10.1002/jgm.3726. J Gene Med. 2024. PMID: 39160647
-
Efficient Non-Viral T-Cell Engineering by Sleeping Beauty Minicircles Diminishing DNA Toxicity and miRNAs Silencing the Endogenous T-Cell Receptors.Hum Gene Ther. 2018 May;29(5):569-584. doi: 10.1089/hum.2017.136. Hum Gene Ther. 2018. PMID: 29562762
-
Generation of CAR+ T Lymphocytes Using the Sleeping Beauty Transposon System.Methods Mol Biol. 2020;2086:131-137. doi: 10.1007/978-1-0716-0146-4_9. Methods Mol Biol. 2020. PMID: 31707672
-
Minicircle-Based Engineering of Chimeric Antigen Receptor (CAR) T Cells.Recent Results Cancer Res. 2016;209:37-50. doi: 10.1007/978-3-319-42934-2_3. Recent Results Cancer Res. 2016. PMID: 28101686 Review.
Cited by
-
Manufacture of T cells using the Sleeping Beauty system to enforce expression of a CD19-specific chimeric antigen receptor.Cancer Gene Ther. 2015 Mar;22(2):95-100. doi: 10.1038/cgt.2014.69. Epub 2015 Jan 16. Cancer Gene Ther. 2015. PMID: 25591810 Review.
-
Enhanced CAR T-cell engineering using non-viral Sleeping Beauty transposition from minicircle vectors.Leukemia. 2017 Jan;31(1):186-194. doi: 10.1038/leu.2016.180. Epub 2016 Jun 24. Leukemia. 2017. PMID: 27491640
-
Creation of versatile cloning platforms for transgene expression and dCas9-based epigenome editing.Nucleic Acids Res. 2019 Feb 28;47(4):e23. doi: 10.1093/nar/gky1286. Nucleic Acids Res. 2019. PMID: 30590691 Free PMC article.
-
CAR T-Cell Production Using Nonviral Approaches.J Immunol Res. 2021 Mar 27;2021:6644685. doi: 10.1155/2021/6644685. eCollection 2021. J Immunol Res. 2021. PMID: 33855089 Free PMC article. Review.
-
Efficient lentiviral transduction method to gene modify cord blood CD8+ T cells for cancer therapy applications.Mol Ther Methods Clin Dev. 2021 Mar 23;21:357-368. doi: 10.1016/j.omtm.2021.03.015. eCollection 2021 Jun 11. Mol Ther Methods Clin Dev. 2021. PMID: 33898633 Free PMC article.
References
-
- Jena B, Dotti G, Cooper LJ (2010) Redirecting T-cell specificity by introducing a tumor-specific chimeric antigen receptor. Blood 116: 1035–1044 blood-2010-01-043737 [pii];10.1182/blood-2010-01-043737 [doi] - DOI - PMC - PubMed
-
- Hacein-Bey-Abina S, Von Kalle C, Schmidt M, Le Deist F, Wulffraat N, et al. (2003) A serious adverse event after successful gene therapy for X-linked severe combined immunodeficiency. N Engl J Med 348: 255–256. - PubMed
-
- Ott MG, Schmidt M, Schwarzwaelder K, Stein S, Siler U, et al. (2006) Correction of X-linked chronic granulomatous disease by gene therapy, augmented by insertional activation of MDS1-EVI1, PRDM16 or SETBP1. Nat Med 12: 401–409. - PubMed
-
- Scholler J, Brady TL, Binder-Scholl G, Hwang WT, Plesa G, et al. (2012) Decade-long safety and function of retroviral-modified chimeric antigen receptor T cells. Sci Transl Med 4: 132ra53 4/132/132ra53 [pii];10.1126/scitranslmed.3003761 [doi] - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

