RNA-Seq Differentiates Tumour and Host mRNA Expression Changes Induced by Treatment of Human Tumour Xenografts with the VEGFR Tyrosine Kinase Inhibitor Cediranib
- PMID: 23840389
- PMCID: PMC3686868
- DOI: 10.1371/journal.pone.0066003
RNA-Seq Differentiates Tumour and Host mRNA Expression Changes Induced by Treatment of Human Tumour Xenografts with the VEGFR Tyrosine Kinase Inhibitor Cediranib
Abstract
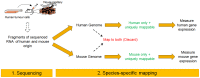
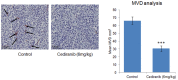
Pre-clinical models of tumour biology often rely on propagating human tumour cells in a mouse. In order to gain insight into the alignment of these models to human disease segments or investigate the effects of different therapeutics, approaches such as PCR or array based expression profiling are often employed despite suffering from biased transcript coverage, and a requirement for specialist experimental protocols to separate tumour and host signals. Here, we describe a computational strategy to profile transcript expression in both the tumour and host compartments of pre-clinical xenograft models from the same RNA sample using RNA-Seq. Key to this strategy is a species-specific mapping approach that removes the need for manipulation of the RNA population, customised sequencing protocols, or prior knowledge of the species component ratio. The method demonstrates comparable performance to species-specific RT-qPCR and a standard microarray platform, and allowed us to quantify gene expression changes in both the tumour and host tissue following treatment with cediranib, a potent vascular endothelial growth factor receptor tyrosine kinase inhibitor, including the reduction of multiple murine transcripts associated with endothelium or vessels, and an increase in genes associated with the inflammatory response in response to cediranib. In the human compartment, we observed a robust induction of hypoxia genes and a reduction in cell cycle associated transcripts. In conclusion, the study establishes that RNA-Seq can be applied to pre-clinical models to gain deeper understanding of model characteristics and compound mechanism of action, and to identify both tumour and host biomarkers.
Conflict of interest statement
Figures

Similar articles
-
Acute vascular response to cediranib treatment in human non-small-cell lung cancer xenografts with different tumour stromal architecture.Lung Cancer. 2015 Nov;90(2):191-8. doi: 10.1016/j.lungcan.2015.08.009. Epub 2015 Aug 20. Lung Cancer. 2015. PMID: 26323213 Free PMC article.
-
Combined MEK and VEGFR inhibition in orthotopic human lung cancer models results in enhanced inhibition of tumor angiogenesis, growth, and metastasis.Clin Cancer Res. 2012 Mar 15;18(6):1641-54. doi: 10.1158/1078-0432.CCR-11-2324. Epub 2012 Jan 24. Clin Cancer Res. 2012. PMID: 22275507 Free PMC article.
-
The HGF/c-MET Pathway Is a Driver and Biomarker of VEGFR-inhibitor Resistance and Vascular Remodeling in Non-Small Cell Lung Cancer.Clin Cancer Res. 2017 Sep 15;23(18):5489-5501. doi: 10.1158/1078-0432.CCR-16-3216. Epub 2017 May 30. Clin Cancer Res. 2017. PMID: 28559461 Free PMC article.
-
Implications of MicroRNAs in the Treatment of Gefitinib-Resistant Non-Small Cell Lung Cancer.Int J Mol Sci. 2016 Feb 15;17(2):237. doi: 10.3390/ijms17020237. Int J Mol Sci. 2016. PMID: 26891293 Free PMC article. Review.
-
Intracranial meningiomas, the VEGF-A pathway, and peritumoral brain oedema.Dan Med J. 2013 Apr;60(4):B4626. Dan Med J. 2013. PMID: 23651727 Review.
Cited by
-
Whole transcriptome profiling of patient-derived xenograft models as a tool to identify both tumor and stromal specific biomarkers.Oncotarget. 2016 Apr 12;7(15):20773-87. doi: 10.18632/oncotarget.8014. Oncotarget. 2016. PMID: 26980748 Free PMC article.
-
Effective combination therapies in preclinical endocrine resistant breast cancer models harboring ER mutations.Oncotarget. 2016 Aug 23;7(34):54120-54136. doi: 10.18632/oncotarget.10852. Oncotarget. 2016. PMID: 27472462 Free PMC article.
-
Tracking cellular and molecular changes in a species-specific manner during experimental tumor progression in vivo.Oncotarget. 2018 Mar 1;9(22):16149-16162. doi: 10.18632/oncotarget.24598. eCollection 2018 Mar 23. Oncotarget. 2018. PMID: 29662633 Free PMC article.
-
Consensus Analysis of Whole Transcriptome Profiles from Two Breast Cancer Patient Cohorts Reveals Long Non-Coding RNAs Associated with Intrinsic Subtype and the Tumour Microenvironment.PLoS One. 2016 Sep 29;11(9):e0163238. doi: 10.1371/journal.pone.0163238. eCollection 2016. PLoS One. 2016. PMID: 27685983 Free PMC article.
-
Preclinical models derived from endoscopic ultrasound-guided tissue acquisition for individualized treatment of pancreatic ductal adenocarcinoma.Front Med (Lausanne). 2023 Jan 5;9:934974. doi: 10.3389/fmed.2022.934974. eCollection 2022. Front Med (Lausanne). 2023. PMID: 36687406 Free PMC article. Review.
References
-
- Wedge SR, Kendrew J, Hennequin LF, Valentine PJ, Barry ST, et al. (2005) cediranib: a highly potent, orally bioavailable, vascular endothelial growth factor receptor-2 tyrosine kinase inhibitor for the treatment of cancer. Cancer Res 65: 4389–4400. - PubMed
-
- Creighton CJ, Bromberg-White JL, Misek DE, Monsma DJ, Brichory FM, et al. (2005) Analysis of tumor-host interactions by gene expression profiling of lung adenocarcinoma xenografts identifies genes involved in tumor formation. Mol Cancer Res 3: 119–129. - PubMed
-
- Harrell JC, Dye WW, Harvell DM, Sartorius CA, Horwitz KB (2008) Contaminating cells alter gene signatures in whole organ versus laser capture microdissected tumors: a comparison of experimental breast cancers and their lymph node metastases. Clin Exp Metastasis 25: 81–88. - PubMed
-
- Wilson CL, Sims AH, Howell A, Miller CJ, Clarke R (2006) Effects of oestrogen on gene expression in epithelium and stroma of normal human breast tissue. Endocr Relat Cancer 13: 617–628. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

