Mechanism of HIV-1 RNA dimerization in the central region of the genome and significance for viral evolution
- PMID: 23839990
- PMCID: PMC3745356
- DOI: 10.1074/jbc.M113.477265
Mechanism of HIV-1 RNA dimerization in the central region of the genome and significance for viral evolution
Abstract
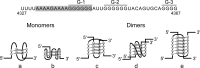
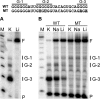
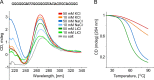
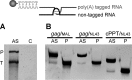
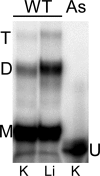
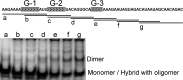
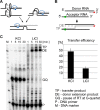
The genome of HIV-1 consists of two identical or nearly identical RNA molecules. The RNA genomes are held in the same, parallel orientation by interactions at the dimer initiation site (DIS). Previous studies showed that in addition to interactions at DIS, sequences located 100 nucleotides downstream from the 5' splice site can dimerize in vitro through an intermolecular G-quartet structure. Here we report that the highly conserved G-rich sequence in the middle portion of the HIV-1 genome near the central polypurine tract (cPPT) dimerizes spontaneously under high ionic strength in the absence of protein. The antisense RNA does not dimerize, strongly indicating that RNA dimerization does not exclusively involve A:U and G:C base pairing. The cation-dependent reverse transcriptase pausing profile, CD spectra profile, and cation-dependent association and thermal dissociation characteristics indicate G-quartet structures. Different forms of G-quartets are formed including monomers and, significantly, intermolecular dimers. Our results indicate that RNA genome dimerization and parallel alignment initiated through interactions at DIS may be greatly expanded and stabilized by formation of an intermolecular G-quartet at a distant site near the cPPT. It is likely that formation of G-quartet structure near the cPPT in vivo keeps the RNA genomes in proximity over a long range, promoting genetic recombination in numerous hot spots.
Keywords: Genome Structure; HIV-1; Homologous Recombination; RNA Structure; Virus.
Figures

Similar articles
-
A short sequence motif in the 5' leader of the HIV-1 genome modulates extended RNA dimer formation and virus replication.J Biol Chem. 2014 Dec 19;289(51):35061-74. doi: 10.1074/jbc.M114.621425. Epub 2014 Nov 3. J Biol Chem. 2014. PMID: 25368321 Free PMC article.
-
A recombination hot spot in HIV-1 contains guanosine runs that can form a G-quartet structure and promote strand transfer in vitro.J Biol Chem. 2009 Dec 4;284(49):33883-93. doi: 10.1074/jbc.M109.055368. Epub 2009 Oct 12. J Biol Chem. 2009. PMID: 19822521 Free PMC article.
-
U3 region in the HIV-1 genome adopts a G-quadruplex structure in its RNA and DNA sequence.Biochemistry. 2014 Apr 29;53(16):2581-93. doi: 10.1021/bi4016692. Epub 2014 Apr 15. Biochemistry. 2014. PMID: 24735378 Free PMC article.
-
A structure-based approach for targeting the HIV-1 genomic RNA dimerization initiation site.Biochimie. 2007 Oct;89(10):1195-203. doi: 10.1016/j.biochi.2007.03.003. Epub 2007 Mar 12. Biochimie. 2007. PMID: 17434658 Review.
-
Dimerization of retroviral genomic RNAs: structural and functional implications.Biochimie. 1996;78(7):639-53. doi: 10.1016/s0300-9084(96)80010-1. Biochimie. 1996. PMID: 8955907 Review.
Cited by
-
G-quadruplexes and G-quadruplex ligands: targets and tools in antiviral therapy.Nucleic Acids Res. 2018 Apr 20;46(7):3270-3283. doi: 10.1093/nar/gky187. Nucleic Acids Res. 2018. PMID: 29554280 Free PMC article. Review.
-
HIV-1 genomic RNA U3 region forms a stable quadruplex-hairpin structure.Biophys Chem. 2021 May;272:106567. doi: 10.1016/j.bpc.2021.106567. Epub 2021 Mar 8. Biophys Chem. 2021. PMID: 33713997 Free PMC article.
-
Colocalization of m6A and G-Quadruplex-Forming Sequences in Viral RNA (HIV, Zika, Hepatitis B, and SV40) Suggests Topological Control of Adenosine N 6-Methylation.ACS Cent Sci. 2019 Feb 27;5(2):218-228. doi: 10.1021/acscentsci.8b00963. Epub 2019 Feb 4. ACS Cent Sci. 2019. PMID: 30834310 Free PMC article. Review.
-
Viral G-quadruplexes: New frontiers in virus pathogenesis and antiviral therapy.Annu Rep Med Chem. 2020;54:101-131. doi: 10.1016/bs.armc.2020.04.001. Epub 2020 May 18. Annu Rep Med Chem. 2020. PMID: 32427223 Free PMC article.
-
G-Quadruplexes in Human Telomere: Structures, Properties, and Applications.Molecules. 2023 Dec 27;29(1):174. doi: 10.3390/molecules29010174. Molecules. 2023. PMID: 38202757 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

