Loss of CARM1 is linked to reduced HuR function in replicative senescence
- PMID: 23837869
- PMCID: PMC3718661
- DOI: 10.1186/1471-2199-14-15
Loss of CARM1 is linked to reduced HuR function in replicative senescence
Abstract
Background: The co-activator-associated arginine methyltransferase 1 (CARM1) catalyzes the methylation of HuR. However, the functional impact of this modification is not fully understood. Here, we investigated the influence of HuR methylation by CARM1 upon the turnover of HuR target mRNAs encoding senescence-regulatory proteins.
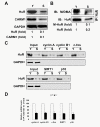
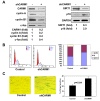
Results: Changing the methylation status of HuR in HeLa cells by either silencing CARM1 or mutating the major methylation site (R217K) greatly diminished the effect of HuR in regulating the turnover of mRNAs encoding cyclin A, cyclin B1, c-fos, SIRT1, and p16. Although knockdown of CARM1 or HuR individually influenced the expression of cyclin A, cyclin B1, c-fos, SIRT1, and p16, joint knockdown of both CARM1 and HuR did not show further effect. Methylation by CARM1 enhanced the association of HuR with the 3'UTR of p16 mRNA, but not with the 3'UTR of cyclin A, cyclin B1, c-fos, or SIRT1 mRNAs. In senescent human diploid fibroblasts (HDFs), reduced CARM1 was accompanied by reduced HuR methylation. In addition, knockdown of CARM1 or mutation of the major methylation site of HuR in HDF markedly impaired the ability of HuR to regulate the expression of cyclin A, cyclin B1, c-fos, SIRT1, and p16 as well to maintain a proliferative phenotype.
Conclusion: CARM1 represses replicative senescence by methylating HuR and thereby enhancing HuR's ability to regulate the turnover of cyclin A, cyclin B1, c-fos, SIRT1, and p16 mRNAs.
Figures




Similar articles
-
Increased AMP:ATP ratio and AMP-activated protein kinase activity during cellular senescence linked to reduced HuR function.J Biol Chem. 2003 Jul 18;278(29):27016-23. doi: 10.1074/jbc.M300318200. Epub 2003 May 1. J Biol Chem. 2003. PMID: 12730239
-
The tRNA methyltransferase NSun2 stabilizes p16INK⁴ mRNA by methylating the 3'-untranslated region of p16.Nat Commun. 2012 Mar 6;3:712. doi: 10.1038/ncomms1692. Nat Commun. 2012. PMID: 22395603 Free PMC article.
-
Lipopolysaccharide-induced methylation of HuR, an mRNA-stabilizing protein, by CARM1. Coactivator-associated arginine methyltransferase.J Biol Chem. 2002 Nov 22;277(47):44623-30. doi: 10.1074/jbc.M206187200. Epub 2002 Sep 16. J Biol Chem. 2002. PMID: 12237300
-
HuR function in disease.Front Biosci (Landmark Ed). 2012 Jan 1;17(1):189-205. doi: 10.2741/3921. Front Biosci (Landmark Ed). 2012. PMID: 22201738 Free PMC article. Review.
-
Functional interplay between RNA-binding protein HuR and microRNAs.Curr Protein Pept Sci. 2012 Jun;13(4):372-9. doi: 10.2174/138920312801619394. Curr Protein Pept Sci. 2012. PMID: 22708488 Free PMC article. Review.
Cited by
-
Protein arginine methyltransferase 4 modulates nitric oxide synthase uncoupling and cerebral blood flow in Alzheimer's disease.J Cell Physiol. 2024 Jun;239(6):e30858. doi: 10.1002/jcp.30858. Epub 2022 Aug 29. J Cell Physiol. 2024. PMID: 36036549
-
PARP1 promotes gene expression at the post-transcriptiona level by modulating the RNA-binding protein HuR.Nat Commun. 2017 Mar 8;8:14632. doi: 10.1038/ncomms14632. Nat Commun. 2017. PMID: 28272405 Free PMC article.
-
RNA-binding protein HuR suppresses senescence through Atg7 mediated autophagy activation in diabetic intervertebral disc degeneration.Cell Prolif. 2021 Feb;54(2):e12975. doi: 10.1111/cpr.12975. Epub 2020 Dec 28. Cell Prolif. 2021. PMID: 33372336 Free PMC article.
-
Hu Antigen R (HuR) Protein Structure, Function and Regulation in Hepatobiliary Tumors.Cancers (Basel). 2022 May 27;14(11):2666. doi: 10.3390/cancers14112666. Cancers (Basel). 2022. PMID: 35681645 Free PMC article. Review.
-
PRMT7 methylates and suppresses GLI2 binding to SUFU thereby promoting its activation.Cell Death Differ. 2020 Jan;27(1):15-28. doi: 10.1038/s41418-019-0334-5. Epub 2019 Apr 18. Cell Death Differ. 2020. PMID: 31000813 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

