Interaction between 25S rRNA A loop and eukaryotic translation initiation factor 5B promotes subunit joining and ensures stringent AUG selection
- PMID: 23836883
- PMCID: PMC3753867
- DOI: 10.1128/MCB.00771-13
Interaction between 25S rRNA A loop and eukaryotic translation initiation factor 5B promotes subunit joining and ensures stringent AUG selection
Abstract
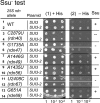
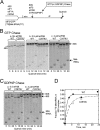
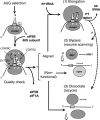
In yeast, 25S rRNA makes up the major mass and shape of the 60S ribosomal subunit. During the last step of translation initiation, eukaryotic initiation factor 5B (eIF5B) promotes the 60S subunit joining with the 40S initiation complex (IC). Malfunctional 60S subunits produced by misfolding or mutation may disrupt the 40S IC stalling on the start codon, thereby altering the stringency of initiation. Using several point mutations isolated by random mutagenesis, here we studied the role of 25S rRNA in start codon selection. Three mutations changing bases near the ribosome surface had strong effects, allowing the initiating ribosomes to skip both AUG and non-AUG codons: C2879U and U2408C, altering the A loop and P loop, respectively, of the peptidyl transferase center, and G1735A, mapping near a Eukarya-specific bridge to the 40S subunit. Overexpression of eIF5B specifically suppressed the phenotype caused by C2879U, suggesting functional interaction between eIF5B and the A loop. In vitro reconstitution assays showed that C2879U decreased eIF5B-catalyzed 60S subunit joining with a 40S IC. Thus, eIF5B interaction with the peptidyl transferase center A loop increases the accuracy of initiation by stabilizing the overall conformation of the 80S initiation complex. This study provides an insight into the effect of ribosomal mutations on translation profiles in eukaryotes.
Figures



Similar articles
-
The Interaction between the Ribosomal Stalk Proteins and Translation Initiation Factor 5B Promotes Translation Initiation.Mol Cell Biol. 2018 Jul 30;38(16):e00067-18. doi: 10.1128/MCB.00067-18. Print 2018 Aug 15. Mol Cell Biol. 2018. PMID: 29844065 Free PMC article.
-
β-Hairpin loop of eukaryotic initiation factor 1 (eIF1) mediates 40 S ribosome binding to regulate initiator tRNA(Met) recruitment and accuracy of AUG selection in vivo.J Biol Chem. 2013 Sep 20;288(38):27546-27562. doi: 10.1074/jbc.M113.498642. Epub 2013 Jul 26. J Biol Chem. 2013. PMID: 23893413 Free PMC article.
-
Proofreading of pre-40S ribosome maturation by a translation initiation factor and 60S subunits.Nat Struct Mol Biol. 2012 Aug;19(8):744-53. doi: 10.1038/nsmb.2308. Epub 2012 Jul 1. Nat Struct Mol Biol. 2012. PMID: 22751017 Free PMC article.
-
Structural Insights into the Mechanism of Scanning and Start Codon Recognition in Eukaryotic Translation Initiation.Trends Biochem Sci. 2017 Aug;42(8):589-611. doi: 10.1016/j.tibs.2017.03.004. Epub 2017 Apr 22. Trends Biochem Sci. 2017. PMID: 28442192 Review.
-
IRES-induced conformational changes in the ribosome and the mechanism of translation initiation by internal ribosomal entry.Biochim Biophys Acta. 2009 Sep-Oct;1789(9-10):558-70. doi: 10.1016/j.bbagrm.2009.06.001. Epub 2009 Jun 17. Biochim Biophys Acta. 2009. PMID: 19539793 Free PMC article. Review.
Cited by
-
Structure of the mammalian 80S initiation complex with initiation factor 5B on HCV-IRES RNA.Nat Struct Mol Biol. 2014 Aug;21(8):721-7. doi: 10.1038/nsmb.2859. Epub 2014 Jul 27. Nat Struct Mol Biol. 2014. PMID: 25064512
-
Sliding of a 43S ribosomal complex from the recognized AUG codon triggered by a delay in eIF2-bound GTP hydrolysis.Nucleic Acids Res. 2016 Feb 29;44(4):1882-93. doi: 10.1093/nar/gkv1514. Epub 2015 Dec 29. Nucleic Acids Res. 2016. PMID: 26717981 Free PMC article.
-
Why is start codon selection so precise in eukaryotes?Translation (Austin). 2014 Mar 12;2(1):e28387. doi: 10.4161/trla.28387. eCollection 2014. Translation (Austin). 2014. PMID: 26779403 Free PMC article. Review.
-
Homoiterons and expansion in ribosomal RNAs.FEBS Open Bio. 2015 Oct 23;5:864-76. doi: 10.1016/j.fob.2015.10.005. eCollection 2015. FEBS Open Bio. 2015. PMID: 26636029 Free PMC article.
-
Wide mutational analysis to ascertain the functional roles of eL33 in ribosome biogenesis and translation initiation.Curr Genet. 2022 Dec;68(5-6):619-644. doi: 10.1007/s00294-022-01251-1. Epub 2022 Aug 22. Curr Genet. 2022. PMID: 35994100 Free PMC article.
References
-
- Algire MA, Maag D, Lorsch JR. 2005. Pi release from eIF2, not GTP hydrolysis, is the step controlled by start-site selection during eukaryotic translation initiation. Mol. Cell 20:251–262 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
