Radical SAM-mediated methylation reactions
- PMID: 23835516
- PMCID: PMC3799849
- DOI: 10.1016/j.cbpa.2013.05.032
Radical SAM-mediated methylation reactions
Abstract
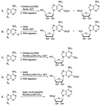
A subset of enzymes that belong to the radical S-adenosylmethionine (SAM) superfamily is able to catalyze methylation reactions. Substrates of these enzymes are distinct from the nucleophilic substrates that undergo methylation by a polar mechanism. Recently, activities of several radical SAM methylating enzymes have been reconstituted in vitro and their mechanisms of catalysis investigated. The RNA modifying enzymes RlmN and Cfr catalyze methylation via a methyl synthase mechanism. These enzymes use SAM in two distinct roles: as a source of a methyl group transferred to a conserved cysteine and as a source of 5'-deoxyadenosyl radical (5'-dA). Hydrogen atom abstraction by this species generates a thiomethylene radical which adds into the RNA substrate, forming an enzyme-substrate covalent adduct. In another recent study, methylation of the indole moiety of tryptophan by the radical SAM and cobalamin-binding domain enzyme TsrM has been reconstituted. Methylcobalamin serves as an intermediate methyl donor in TsrM, and is proposed to transfer the methyl group as a methyl radical. Interestingly, despite the presence of the radical SAM motif, no reductive cleavage of SAM has been observed in this methylation. These important reconstitutions set the stage for further studies on mechanisms of radical methylation.
Copyright © 2013 Elsevier Ltd. All rights reserved.
Figures


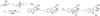
Similar articles
-
Radical-mediated enzymatic methylation: a tale of two SAMS.Acc Chem Res. 2012 Apr 17;45(4):555-64. doi: 10.1021/ar200202c. Epub 2011 Nov 18. Acc Chem Res. 2012. PMID: 22097883 Free PMC article. Review.
-
Efficient methylation of C2 in l-tryptophan by the cobalamin-dependent radical S-adenosylmethionine methylase TsrM requires an unmodified N1 amine.J Biol Chem. 2017 Sep 15;292(37):15456-15467. doi: 10.1074/jbc.M117.778548. Epub 2017 Jul 26. J Biol Chem. 2017. PMID: 28747433 Free PMC article.
-
Understanding the role of electron donors in the reaction catalyzed by Tsrm, a cobalamin-dependent radical S-adenosylmethionine methylase.J Biol Inorg Chem. 2019 Sep;24(6):831-839. doi: 10.1007/s00775-019-01689-8. Epub 2019 Jul 26. J Biol Inorg Chem. 2019. PMID: 31350635 Free PMC article.
-
Structural basis for non-radical catalysis by TsrM, a radical SAM methylase.Nat Chem Biol. 2021 Apr;17(4):485-491. doi: 10.1038/s41589-020-00717-y. Epub 2021 Jan 18. Nat Chem Biol. 2021. PMID: 33462497 Free PMC article.
-
Mechanistic diversity of radical S-adenosylmethionine (SAM)-dependent methylation.J Biol Chem. 2015 Feb 13;290(7):3995-4002. doi: 10.1074/jbc.R114.607044. Epub 2014 Dec 4. J Biol Chem. 2015. PMID: 25477520 Free PMC article. Review.
Cited by
-
Methyltransferases: Functions and Applications.Chembiochem. 2022 Sep 16;23(18):e202200212. doi: 10.1002/cbic.202200212. Epub 2022 Jul 5. Chembiochem. 2022. PMID: 35691829 Free PMC article. Review.
-
C-H methylation of heteroarenes inspired by radical SAM methyl transferase.J Am Chem Soc. 2014 Apr 2;136(13):4853-6. doi: 10.1021/ja5007838. Epub 2014 Mar 21. J Am Chem Soc. 2014. PMID: 24611732 Free PMC article.
-
The Hydrogenobyric Acid Structure Reveals the Corrin Ligand as an Entatic State Module Empowering B12 Cofactors for Catalysis.Angew Chem Int Ed Engl. 2019 Jul 29;58(31):10756-10760. doi: 10.1002/anie.201904713. Epub 2019 Jun 26. Angew Chem Int Ed Engl. 2019. PMID: 31115943 Free PMC article.
-
Radical S-Adenosylmethionine Enzymes Involved in RiPP Biosynthesis.Biochemistry. 2017 Oct 10;56(40):5229-5244. doi: 10.1021/acs.biochem.7b00771. Epub 2017 Sep 22. Biochemistry. 2017. PMID: 28895719 Free PMC article. Review.
-
Replacement of the Cobalt Center of Vitamin B12 by Nickel: Nibalamin and Nibyric Acid Prepared from Metal-Free B12 Ligands Hydrogenobalamin and Hydrogenobyric Acid.Angew Chem Int Ed Engl. 2020 Nov 2;59(45):20129-20136. doi: 10.1002/anie.202008407. Epub 2020 Sep 2. Angew Chem Int Ed Engl. 2020. PMID: 32686888 Free PMC article.
References
-
- Sofia HJ, Chen G, Hetzler BG, Reyes-Spindola JF, Miller NE. Radical SAM, a novel protein superfamily linking unresolved steps in familiar biosynthetic pathways with radical mechanisms: functional characterization using new analysis and information visualization methods. Nucleic Acids Res. 2001;29:1097–1106. - PMC - PubMed
-
- Frey PA, Magnusson OT. S-Adenosylmethionine: a wolf in sheep's clothing, or a rich man's adenosylcobalamin? Chem Rev. 2003;103:2129–2148. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

