Evolutionary origins, molecular cloning and expression of carotenoid hydroxylases in eukaryotic photosynthetic algae
- PMID: 23834441
- PMCID: PMC3728230
- DOI: 10.1186/1471-2164-14-457
Evolutionary origins, molecular cloning and expression of carotenoid hydroxylases in eukaryotic photosynthetic algae
Abstract
Background: Xanthophylls, oxygenated derivatives of carotenes, play critical roles in photosynthetic apparatus of cyanobacteria, algae, and higher plants. Although the xanthophylls biosynthetic pathway of algae is largely unknown, it is of particular interest because they have a very complicated evolutionary history. Carotenoid hydroxylase (CHY) is an important protein that plays essential roles in xanthophylls biosynthesis. With the availability of 18 sequenced algal genomes, we performed a comprehensive comparative analysis of chy genes and explored their distribution, structure, evolution, origins, and expression.
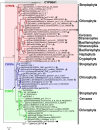
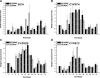
Results: Overall 60 putative chy genes were identified and classified into two major subfamilies (bch and cyp97) according to their domain structures. Genes in the bch subfamily were found in 10 green algae and 1 red alga, but absent in other algae. In the phylogenetic tree, bch genes of green algae and higher plants share a common ancestor and are of non-cyanobacterial origin, whereas that of red algae is of cyanobacteria. The homologs of cyp97a/c genes were widespread only in green algae, while cyp97b paralogs were seen in most of algae. Phylogenetic analysis on cyp97 genes supported the hypothesis that cyp97b is an ancient gene originated before the formation of extant algal groups. The cyp97a gene is more closely related to cyp97c in evolution than to cyp97b. The two cyp97 genes were isolated from the green alga Haematococcus pluvialis, and transcriptional expression profiles of chy genes were observed under high light stress of different wavelength.
Conclusions: Green algae received a β-xanthophylls biosynthetic pathway from host organisms. Although red algae inherited the pathway from cyanobacteria during primary endosymbiosis, it remains unclear in Chromalveolates. The α-xanthophylls biosynthetic pathway is a common feature in green algae and higher plants. The origination of cyp97a/c is most likely due to gene duplication before divergence of green algae and higher plants. Protein domain structures and expression analyses in green alga H. pluvialis indicate that various chy genes are in different manners response to light. The knowledge of evolution of chy genes in photosynthetic eukaryotes provided information of gene cloning and functional investigation of chy genes in algae in the future.
Figures




Similar articles
-
Occurrence and environmental stress responses of two plastid terminal oxidases in Haematococcus pluvialis (Chlorophyceae).Planta. 2009 Jun;230(1):191-203. doi: 10.1007/s00425-009-0932-4. Epub 2009 May 1. Planta. 2009. PMID: 19408010
-
Light induction of carotenoid biosynthesis genes in the green alga Haematococcus pluvialis: regulation by photosynthetic redox control.Plant Mol Biol. 2003 May;52(2):343-56. doi: 10.1023/a:1023948929665. Plant Mol Biol. 2003. PMID: 12856941
-
Carotenoid biosynthesis in the primitive red alga Cyanidioschyzon merolae.Eukaryot Cell. 2007 Mar;6(3):533-45. doi: 10.1128/EC.00265-06. Epub 2006 Nov 3. Eukaryot Cell. 2007. PMID: 17085635 Free PMC article.
-
What was the real contribution of endosymbionts to the eukaryotic nucleus? Insights from photosynthetic eukaryotes.Cold Spring Harb Perspect Biol. 2014 Jul 1;6(7):a016014. doi: 10.1101/cshperspect.a016014. Cold Spring Harb Perspect Biol. 2014. PMID: 24984774 Free PMC article. Review.
-
Applications of next-generation sequencing to unravelling the evolutionary history of algae.Int J Syst Evol Microbiol. 2014 Feb;64(Pt 2):333-345. doi: 10.1099/ijs.0.054221-0. Int J Syst Evol Microbiol. 2014. PMID: 24505071 Review.
Cited by
-
Biosynthetic routes of hydroxylated carotenoids (xanthophylls) in Marchantia polymorpha, and production of novel and rare xanthophylls through pathway engineering in Escherichia coli.Planta. 2015 Mar;241(3):699-710. doi: 10.1007/s00425-014-2213-0. Epub 2014 Dec 3. Planta. 2015. PMID: 25467956
-
Cytochromes P450 evolution in the plant terrestrialization context.Philos Trans R Soc Lond B Biol Sci. 2024 Nov 18;379(1914):20230363. doi: 10.1098/rstb.2023.0363. Epub 2024 Sep 30. Philos Trans R Soc Lond B Biol Sci. 2024. PMID: 39343021 Review.
-
Genome-Scale Metabolic Networks Shed Light on the Carotenoid Biosynthesis Pathway in the Brown Algae Saccharina japonica and Cladosiphon okamuranus.Antioxidants (Basel). 2019 Nov 16;8(11):564. doi: 10.3390/antiox8110564. Antioxidants (Basel). 2019. PMID: 31744163 Free PMC article.
-
Cytochrome P450 CitCYP97B modulates carotenoid accumulation diversity by hydroxylating β-cryptoxanthin in Citrus.Plant Commun. 2024 Jun 10;5(6):100847. doi: 10.1016/j.xplc.2024.100847. Epub 2024 Feb 19. Plant Commun. 2024. PMID: 38379285 Free PMC article.
-
Synthetic biology and metabolic engineering for marine carotenoids: new opportunities and future prospects.Mar Drugs. 2014 Sep 17;12(9):4810-32. doi: 10.3390/md12094810. Mar Drugs. 2014. PMID: 25233369 Free PMC article. Review.
References
-
- Hirschberg J, Cohen M, Harker M, Lotan T, Mann V, Pecker I. Molecular genetics of the carotenoid biosynthesis pathway in plants and algae. Pure Appl Chem. 1997;69(10):2151–2158. doi: 10.1351/pac199769102151. - DOI
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

