Cross-species analysis of genic GC3 content and DNA methylation patterns
- PMID: 23833164
- PMCID: PMC3762193
- DOI: 10.1093/gbe/evt103
Cross-species analysis of genic GC3 content and DNA methylation patterns
Abstract
The GC content in the third codon position (GC(3)) exhibits a unimodal distribution in many plant and animal genomes. Interestingly, grasses and homeotherm vertebrates exhibit a unique bimodal distribution. High GC(3) was previously found to be associated with variable expression, higher frequency of upstream TATA boxes, and an increase of GC(3) from 5' to 3'. Moreover, GC(3)-rich genes are predominant in certain gene classes and are enriched in CpG dinucleotides that are potential targets for methylation. Based on the GC(3) bimodal distribution we hypothesize that GC(3) has a regulatory role involving methylation and gene expression. To test that hypothesis, we selected diverse taxa (rice, thale cress, bee, and human) that varied in the modality of their GC(3) distribution and tested the association between GC(3), DNA methylation, and gene expression. We examine the relationship between cytosine methylation levels and GC(3), gene expression, genome signature, gene length, and other gene compositional features. We find a strong negative correlation (Pearson's correlation coefficient r = -0.67, P value < 0.0001) between GC(3) and genic CpG methylation. The comparison between 5'-3' gradients of CG(3)-skew and genic methylation for the taxa in the study suggests interplay between gene-body methylation and transcription-coupled cytosine deamination effect. Compositional features are correlated with methylation levels of genes in rice, thale cress, human, bee, and fruit fly (which acts as an unmethylated control). These patterns allow us to generate evolutionary hypotheses about the relationships between GC(3) and methylation and how these affect expression patterns. Specifically, we propose that the opposite effects of methylation and compositional gradients along coding regions of GC(3)-poor and GC(3)-rich genes are the products of several competing processes.
Keywords: Apis mellifera; Arabidopsis thaliana; DNA methylation; GC3; Homo sapiens; Oryza sativa; gene expression; grasses; homeotherms.
Figures
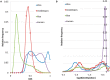
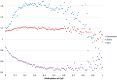
 . Standard error of the mean does not exceed 0.05 for the mean methylation levels calculation.
. Standard error of the mean does not exceed 0.05 for the mean methylation levels calculation.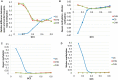
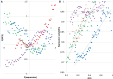
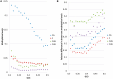
 as a function of GC3 between worker and queen bee. (B) Difference in gene-body methylation between worker and queen bee as a function of GC3. (C) Queen and (D) worker bee methylation as a function of GC3. Every point represents an average of at least 228 genes. Standard error of the mean for methylation levels was below 0.006.
as a function of GC3 between worker and queen bee. (B) Difference in gene-body methylation between worker and queen bee as a function of GC3. (C) Queen and (D) worker bee methylation as a function of GC3. Every point represents an average of at least 228 genes. Standard error of the mean for methylation levels was below 0.006.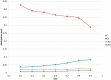
Similar articles
-
Gene expression and nucleotide composition are associated with genic methylation level in Oryza sativa.BMC Bioinformatics. 2014 Jan 21;15:23. doi: 10.1186/1471-2105-15-23. BMC Bioinformatics. 2014. PMID: 24447369 Free PMC article.
-
GC-compositional strand bias around transcription start sites in plants and fungi.BMC Genomics. 2005 Feb 28;6:26. doi: 10.1186/1471-2164-6-26. BMC Genomics. 2005. PMID: 15733327 Free PMC article.
-
The bimodal distribution of genic GC content is ancestral to monocot species.Genome Biol Evol. 2014 Dec 19;7(1):336-48. doi: 10.1093/gbe/evu278. Genome Biol Evol. 2014. PMID: 25527839 Free PMC article.
-
GC content evolution in coding regions of angiosperm genomes: a unifying hypothesis.Trends Genet. 2014 Jul;30(7):263-70. doi: 10.1016/j.tig.2014.05.002. Epub 2014 Jun 7. Trends Genet. 2014. PMID: 24916172 Review.
-
The vertebrate genome: isochores and evolution.Mol Biol Evol. 1993 Jan;10(1):186-204. doi: 10.1093/oxfordjournals.molbev.a039994. Mol Biol Evol. 1993. PMID: 8450755 Review.
Cited by
-
Evolutionary Protection of Krüppel-Like Factors 2 and 4 in the Development of the Mature Hemovascular System.Front Cardiovasc Med. 2021 May 17;8:645719. doi: 10.3389/fcvm.2021.645719. eCollection 2021. Front Cardiovasc Med. 2021. PMID: 34079826 Free PMC article.
-
Analysis of codon usage patterns in citrus based on coding sequence data.BMC Genomics. 2020 Dec 16;21(Suppl 5):234. doi: 10.1186/s12864-020-6641-x. BMC Genomics. 2020. PMID: 33327935 Free PMC article.
-
GC heterogeneity reveals sequence-structures evolution of angiosperm ITS2.BMC Plant Biol. 2023 Dec 1;23(1):608. doi: 10.1186/s12870-023-04634-9. BMC Plant Biol. 2023. PMID: 38036992 Free PMC article.
-
The mysterious orphans of Mycoplasmataceae.Biol Direct. 2016 Jan 8;11(1):2. doi: 10.1186/s13062-015-0104-3. Biol Direct. 2016. PMID: 26747447 Free PMC article.
-
Evolution of Brain Active Gene Promoters in Human Lineage Towards the Increased Plasticity of Gene Regulation.Mol Neurobiol. 2018 Mar;55(3):1871-1904. doi: 10.1007/s12035-017-0427-4. Epub 2017 Feb 24. Mol Neurobiol. 2018. PMID: 28233272
References
-
- Aamodt RM. Age-and caste-dependent decrease in expression of genes maintaining DNA and RNA quality and mitochondrial integrity in the honeybee wing muscle. Exp Gerontol. 2009;44(9):586–593. - PubMed
-
- Adams J. Imprinting and genetic disease: Angelman, Prader-Willi and Beckwith-Weidemann syndromes. Nat Educ. 2008;1(1)
-
- Anastasiadou C, Malousi A, Maglaveras N, Kouidou S. Human epigenome data reveal increased CpG methylation in alternatively spliced sites and putative exonic splicing enhancers. DNA Cell Biol. 2011;30(5):267–275. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

