PTree: pattern-based, stochastic search for maximum parsimony phylogenies
- PMID: 23825794
- PMCID: PMC3698465
- DOI: 10.7717/peerj.89
PTree: pattern-based, stochastic search for maximum parsimony phylogenies
Abstract
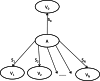
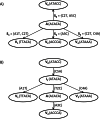
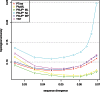
Phylogenetic reconstruction is vital to analyzing the evolutionary relationship of genes within and across populations of different species. Nowadays, with next generation sequencing technologies producing sets comprising thousands of sequences, robust identification of the tree topology, which is optimal according to standard criteria such as maximum parsimony, maximum likelihood or posterior probability, with phylogenetic inference methods is a computationally very demanding task. Here, we describe a stochastic search method for a maximum parsimony tree, implemented in a software package we named PTree. Our method is based on a new pattern-based technique that enables us to infer intermediate sequences efficiently where the incorporation of these sequences in the current tree topology yields a phylogenetic tree with a lower cost. Evaluation across multiple datasets showed that our method is comparable to the algorithms implemented in PAUP* or TNT, which are widely used by the bioinformatics community, in terms of topological accuracy and runtime. We show that our method can process large-scale datasets of 1,000-8,000 sequences. We believe that our novel pattern-based method enriches the current set of tools and methods for phylogenetic tree inference. The software is available under: http://algbio.cs.uni-duesseldorf.de/webapps/wa-download/.
Keywords: Local search; Maximum parsimony; Phylogeny reconstruction; Stochastic search.
Figures
Similar articles
-
MPBoot: fast phylogenetic maximum parsimony tree inference and bootstrap approximation.BMC Evol Biol. 2018 Feb 2;18(1):11. doi: 10.1186/s12862-018-1131-3. BMC Evol Biol. 2018. PMID: 29390973 Free PMC article.
-
Inference of gain and loss events from phyletic patterns using stochastic mapping and maximum parsimony--a simulation study.Genome Biol Evol. 2011;3:1265-75. doi: 10.1093/gbe/evr101. Epub 2011 Oct 4. Genome Biol Evol. 2011. PMID: 21971516 Free PMC article.
-
IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies.Mol Biol Evol. 2015 Jan;32(1):268-74. doi: 10.1093/molbev/msu300. Epub 2014 Nov 3. Mol Biol Evol. 2015. PMID: 25371430 Free PMC article.
-
Stochastic search strategy for estimation of maximum likelihood phylogenetic trees.Syst Biol. 2001 Feb;50(1):7-17. Syst Biol. 2001. PMID: 12116596
-
Towards improving searches for optimal phylogenies.Syst Biol. 2015 Jan;64(1):56-65. doi: 10.1093/sysbio/syu065. Epub 2014 Aug 26. Syst Biol. 2015. PMID: 25164916
References
-
- EuResist 2010. HIV reverse transcriptase and polymerase sequences. Available at http://www.euresist.org/web/guest (accessed 12 April 2010)
-
- Felsenstein J. Cases in which parsimony or compatibility methods will be positively misleading. Systematic Zoology. 1978;27:401–410. doi: 10.2307/2412923. - DOI
-
- Felsenstein J. Inferring phylogenies. 2nd ed. Sunderland: Sinauer Associates; 2003.
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

