A New Method of the Visualization of the Double-Stranded Mitochondrial and Nuclear DNA
- PMID: 23825578
- PMCID: PMC3688954
- DOI: 10.1371/journal.pone.0066864
A New Method of the Visualization of the Double-Stranded Mitochondrial and Nuclear DNA
Abstract
The study describes the method of a sensitive detection of double-stranded DNA molecules in situ. It is based on the oxidative attack on the deoxyribose moiety by copper(I) in the presence of oxygen. We have shown previously that the oxidative attack leads to the formation of frequent gaps in DNA. Here we have demonstrated that the gaps can be utilized as the origins for an efficient synthesis of complementary labeled strands by DNA polymerase I and that such enzymatic detection of the double-stranded DNA is a sensitive approach enabling in-situ detection of both the nuclear and mitochondrial genomes in formaldehyde-fixed human cells.
Conflict of interest statement
Figures
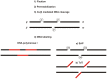
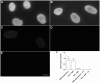

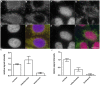
Similar articles
-
Tracking Mitochondrial DNA In Situ.Methods Mol Biol. 2016;1351:81-92. doi: 10.1007/978-1-4939-3040-1_7. Methods Mol Biol. 2016. PMID: 26530676
-
Atomic scissors: a new method of tracking the 5-bromo-2'-deoxyuridine-labeled DNA in situ.PLoS One. 2012;7(12):e52584. doi: 10.1371/journal.pone.0052584. Epub 2012 Dec 26. PLoS One. 2012. PMID: 23300711 Free PMC article.
-
Prevalent coordination of mitochondrial DNA transcription and initiation of replication with the cell cycle.Nucleic Acids Res. 2013 Mar 1;41(5):3068-78. doi: 10.1093/nar/gkt015. Epub 2013 Jan 23. Nucleic Acids Res. 2013. PMID: 23345615 Free PMC article.
-
Nuclear and mitochondrial DNA oxidation in Alzheimer's disease.Free Radic Res. 2012 Apr;46(4):565-76. doi: 10.3109/10715762.2011.648188. Epub 2012 Jan 23. Free Radic Res. 2012. PMID: 22149654 Review.
-
Mitochondria-nucleus network for genome stability.Free Radic Biol Med. 2015 May;82:73-104. doi: 10.1016/j.freeradbiomed.2015.01.013. Epub 2015 Jan 30. Free Radic Biol Med. 2015. PMID: 25640729 Review.
Cited by
-
The Fingerprint of Anti-Bromodeoxyuridine Antibodies and Its Use for the Assessment of Their Affinity to 5-Bromo-2'-Deoxyuridine in Cellular DNA under Various Conditions.PLoS One. 2015 Jul 10;10(7):e0132393. doi: 10.1371/journal.pone.0132393. eCollection 2015. PLoS One. 2015. PMID: 26161977 Free PMC article.
-
A New Sensitive Method for the Detection of Mycoplasmas Using Fluorescence Microscopy.Cells. 2019 Nov 25;8(12):1510. doi: 10.3390/cells8121510. Cells. 2019. PMID: 31775352 Free PMC article.
-
New Concept and Apparatus for Cytocentrifugation and Cell Processing for Microscopy Analysis.Int J Mol Sci. 2021 Jul 1;22(13):7098. doi: 10.3390/ijms22137098. Int J Mol Sci. 2021. PMID: 34281153 Free PMC article.
References
-
- Colson P, Houssier C, Bailly C (1995) Use of electric linear dichroism and competition experiments with intercalating drugs to investigate the mode of binding of Hoechst 33258, berenil and DAPI to GC sequences. J Biomol Struct Dyn 13: 351–366. - PubMed
-
- Kapuscinski J (1995) DAPI: a DNA-specific fluorescent probe. Biotech Histochem 70: 220–233. - PubMed
-
- Hirons GT, Fawcett JJ, Crissman HA (1994) TOTO and YOYO: new very bright fluorochromes for DNA content analyses by flow cytometry. Cytometry 15: 129–140. - PubMed
-
- Tanious FA, Veal JM, Buczak H, Ratmeyer LS, Wilson WD (1992) DAPI (4′,6-diamidino-2-phenylindole) binds differently to DNA and RNA: minor-groove binding at AT sites and intercalation at AU sites. Biochemistry 31: 3103–3112. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

