On homology modeling of the M₂ muscarinic acetylcholine receptor subtype
- PMID: 23812908
- PMCID: PMC3717152
- DOI: 10.1007/s10822-013-9660-8
On homology modeling of the M₂ muscarinic acetylcholine receptor subtype
Abstract
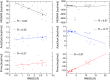
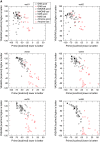
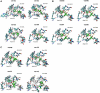
Twelve homology models of the human M2 muscarinic receptor using different sets of templates have been designed using the Prime program or the modeller program and compared to crystallographic structure (PDB:3UON). The best models were obtained using single template of the closest published structure, the M3 muscarinic receptor (PDB:4DAJ). Adding more (structurally distant) templates led to worse models. Data document a key role of the template in homology modeling. The models differ substantially. The quality checks built into the programs do not correlate with the RMSDs to the crystallographic structure and cannot be used to select the best model. Re-docking of the antagonists present in crystallographic structure and relative binding energy estimation by calculating MM/GBSA in Prime and the binding energy function in YASARA suggested it could be possible to evaluate the quality of the orthosteric binding site based on the prediction of relative binding energies. Although estimation of relative binding energies distinguishes between relatively good and bad models it does not indicate the best one. On the other hand, visual inspection of the models for known features and knowledge-based analysis of the intramolecular interactions allows an experimenter to select overall best models manually.
Figures



Similar articles
-
A model of the human M2 muscarinic acetylcholine receptor.J Comput Aided Mol Des. 2002 Nov;16(11):795-801. doi: 10.1023/a:1023880611709. J Comput Aided Mol Des. 2002. PMID: 12825791
-
Structure of the human M2 muscarinic acetylcholine receptor bound to an antagonist.Nature. 2012 Jan 25;482(7386):547-51. doi: 10.1038/nature10753. Nature. 2012. PMID: 22278061 Free PMC article.
-
Exploring the structure of opioid receptors with homology modeling based on single and multiple templates and subsequent docking: a comparative study.J Mol Model. 2011 May;17(5):1207-21. doi: 10.1007/s00894-010-0803-8. Epub 2010 Jul 27. J Mol Model. 2011. PMID: 20661609
-
Regulation of signal transduction at M2 muscarinic receptor.Physiol Res. 2004;53 Suppl 1:S131-40. Physiol Res. 2004. PMID: 15119944 Review.
-
Dualsteric GPCR targeting and functional selectivity: the paradigmatic M(2) muscarinic acetylcholine receptor.Drug Discov Today Technol. 2013 Summer;10(2):e245-52. doi: 10.1016/j.ddtec.2012.12.003. Drug Discov Today Technol. 2013. PMID: 24050275 Review.
Cited by
-
Lead discovery and in silico 3D structure modeling of tumorigenic FAM72A (p17).Tumour Biol. 2015 Jan;36(1):239-49. doi: 10.1007/s13277-014-2620-7. Epub 2014 Sep 20. Tumour Biol. 2015. PMID: 25234718
-
Discovery of Histone Deacetylase Inhibitor Using Molecular Modeling and Free Energy Calculations.ACS Omega. 2022 May 24;7(22):18786-18794. doi: 10.1021/acsomega.2c01572. eCollection 2022 Jun 7. ACS Omega. 2022. PMID: 35694501 Free PMC article.
-
In silico Prediction, Characterization, Molecular Docking, and Dynamic Studies on Fungal SDRs as Novel Targets for Searching Potential Fungicides Against Fusarium Wilt in Tomato.Front Pharmacol. 2018 Oct 22;9:1038. doi: 10.3389/fphar.2018.01038. eCollection 2018. Front Pharmacol. 2018. PMID: 30405403 Free PMC article.
-
Identification of antiviral phytochemicals as a potential SARS-CoV-2 main protease (Mpro) inhibitor using docking and molecular dynamics simulations.Sci Rep. 2021 Oct 13;11(1):20295. doi: 10.1038/s41598-021-99165-4. Sci Rep. 2021. PMID: 34645849 Free PMC article.
-
Characterization of Plant-Derived Natural Inhibitors of Dipeptidyl Peptidase-4 as Potential Antidiabetic Agents: A Computational Study.Pharmaceutics. 2024 Apr 1;16(4):483. doi: 10.3390/pharmaceutics16040483. Pharmaceutics. 2024. PMID: 38675143 Free PMC article.
References
-
- Jakubík J, Doležal V, El-Fakahany EE, Janíčková H, Randáková A, Šantrůčková E. Perspective for design of selective muscarinic agonists. In: Babušíková E, Dobrota D, Lehotský J, editors. New frontiers in molecular mechanisms in psychiatric and neurologic disorders. Martin: Jesseniova lekárska fakulta UK; 2011.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

