Chromatin modifications as determinants of muscle stem cell quiescence and chronological aging
- PMID: 23810552
- PMCID: PMC4103025
- DOI: 10.1016/j.celrep.2013.05.043
Chromatin modifications as determinants of muscle stem cell quiescence and chronological aging
Abstract
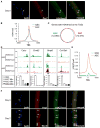
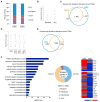
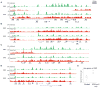
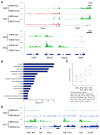
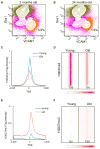
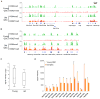
The ability to maintain quiescence is critical for the long-term maintenance of a functional stem cell pool. To date, the epigenetic and transcriptional characteristics of quiescent stem cells and how they change with age remain largely unknown. In this study, we explore the chromatin features of adult skeletal muscle stem cells, or satellite cells (SCs), which reside predominantly in a quiescent state in fully developed limb muscles of both young and aged mice. Using a ChIP-seq approach to obtain global epigenetic profiles of quiescent SCs (QSCs), we show that QSCs possess a permissive chromatin state in which few genes are epigenetically repressed by Polycomb group (PcG)-mediated histone 3 lysine 27 trimethylation (H3K27me3), and a large number of genes encoding regulators that specify nonmyogenic lineages are demarcated by bivalent domains at their transcription start sites (TSSs). By comparing epigenetic profiles of QSCs from young and old mice, we also provide direct evidence that, with age, epigenetic changes accumulate and may lead to a functional decline in quiescent stem cells. These findings highlight the importance of chromatin mapping in understanding unique features of stem cell identity and stem cell aging.
Copyright © 2013 The Authors. Published by Elsevier Inc. All rights reserved.
Figures

Similar articles
-
Genome-wide maps of histone modifications unwind in vivo chromatin states of the hair follicle lineage.Cell Stem Cell. 2011 Sep 2;9(3):219-32. doi: 10.1016/j.stem.2011.07.015. Cell Stem Cell. 2011. PMID: 21885018 Free PMC article.
-
Epigenetic regulation of stem cells : the role of chromatin in cell differentiation.Adv Exp Med Biol. 2013;786:307-28. doi: 10.1007/978-94-007-6621-1_17. Adv Exp Med Biol. 2013. PMID: 23696364
-
Genome-Wide Identification of Transcription Factor-Binding Sites in Quiescent Adult Neural Stem Cells.Methods Mol Biol. 2018;1686:265-286. doi: 10.1007/978-1-4939-7371-2_19. Methods Mol Biol. 2018. PMID: 29030827 Free PMC article.
-
Polycomb group-mediated histone H2A monoubiquitination in epigenome regulation and nuclear processes.Nat Commun. 2020 Nov 23;11(1):5947. doi: 10.1038/s41467-020-19722-9. Nat Commun. 2020. PMID: 33230107 Free PMC article. Review.
-
Epigenetic regulation of T cells by Polycomb group proteins.J Leukoc Biol. 2022 Jun;111(6):1253-1267. doi: 10.1002/JLB.2RI0122-039R. Epub 2022 Apr 25. J Leukoc Biol. 2022. PMID: 35466423 Review.
Cited by
-
Contribution of muscle satellite cells to sarcopenia.Front Physiol. 2022 Aug 12;13:892749. doi: 10.3389/fphys.2022.892749. eCollection 2022. Front Physiol. 2022. PMID: 36035464 Free PMC article. Review.
-
The effect of age on stem cell function and utility for therapy.Cell Med. 2018 Jun 8;10:2155179018773756. doi: 10.1177/2155179018773756. eCollection 2018. Cell Med. 2018. PMID: 32634187 Free PMC article. Review.
-
HIRA stabilizes skeletal muscle lineage identity.Nat Commun. 2021 Jun 8;12(1):3450. doi: 10.1038/s41467-021-23775-9. Nat Commun. 2021. PMID: 34103504 Free PMC article.
-
The epigenome in Alzheimer's disease: current state and approaches for a new path to gene discovery and understanding disease mechanism.Acta Neuropathol. 2016 Oct;132(4):503-14. doi: 10.1007/s00401-016-1612-7. Epub 2016 Aug 29. Acta Neuropathol. 2016. PMID: 27573688 Free PMC article. Review.
-
The Information Theory of Aging.Nat Aging. 2023 Dec;3(12):1486-1499. doi: 10.1038/s43587-023-00527-6. Epub 2023 Dec 15. Nat Aging. 2023. PMID: 38102202 Review.
References
-
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. - PubMed
-
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

