Measles virus nonstructural C protein modulates viral RNA polymerase activity by interacting with host protein SHCBP1
- PMID: 23804634
- PMCID: PMC3754092
- DOI: 10.1128/JVI.00714-13
Measles virus nonstructural C protein modulates viral RNA polymerase activity by interacting with host protein SHCBP1
Abstract
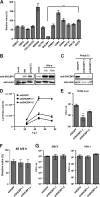
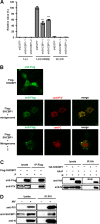
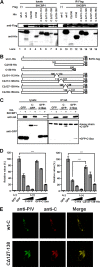
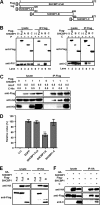
Most viruses possess strategies to circumvent host immune responses. The measles virus (MV) nonstructural C protein suppresses the interferon response, thereby allowing efficient viral growth, but its detailed mechanism has been unknown. We identified Shc Src homology 2 domain-binding protein 1 (SHCBP1) as one of the host proteins interacting with the C protein. Knockdown of SHCBP1 using a short-hairpin RNA greatly reduced MV growth. SHCBP1 was found to be required for viral RNA synthesis in the minigenome assay and to bind to the MV phosphoprotein, a subunit of the viral RNA polymerase. A stretch of 12 amino acid residues in the C protein were sufficient for SHCBP1 binding, and the peptide containing these 12 residues could suppress MV RNA synthesis, like the full-length C protein. The central region of SHCBP1 was found to bind to the C protein, as well as the phosphoprotein, but the two viral proteins did not compete for SHCBP1 binding. Our results indicate that the C protein modulates MV RNA polymerase activity by binding to the host protein SHCBP1. SHCBP1 may be exploited as a target of antiviral compounds.
Figures
Similar articles
-
V and C proteins of measles virus function as virulence factors in vivo.Virology. 2000 Feb 1;267(1):80-9. doi: 10.1006/viro.1999.0118. Virology. 2000. PMID: 10648185
-
Actin-Modulating Protein Cofilin Is Involved in the Formation of Measles Virus Ribonucleoprotein Complex at the Perinuclear Region.J Virol. 2015 Oct;89(20):10524-31. doi: 10.1128/JVI.01819-15. Epub 2015 Aug 12. J Virol. 2015. PMID: 26269174 Free PMC article.
-
Nonstructural C protein is required for efficient measles virus replication in human peripheral blood cells.J Virol. 1999 Feb;73(2):1695-8. doi: 10.1128/JVI.73.2.1695-1698.1999. J Virol. 1999. PMID: 9882382 Free PMC article.
-
The Role of Shcbp1 in Signaling and Disease.Curr Cancer Drug Targets. 2019;19(11):854-862. doi: 10.2174/1568009619666190620114928. Curr Cancer Drug Targets. 2019. PMID: 31250756 Review.
-
Biological functions and therapeutic potential of SHCBP1 in human cancer.Biomed Pharmacother. 2023 Apr;160:114362. doi: 10.1016/j.biopha.2023.114362. Epub 2023 Feb 3. Biomed Pharmacother. 2023. PMID: 36739763 Review.
Cited by
-
The C Protein Is Recruited to Measles Virus Ribonucleocapsids by the Phosphoprotein.J Virol. 2020 Jan 31;94(4):e01733-19. doi: 10.1128/JVI.01733-19. Print 2020 Jan 31. J Virol. 2020. PMID: 31748390 Free PMC article.
-
Evolution and structural organization of the C proteins of paramyxovirinae.PLoS One. 2014 Feb 25;9(2):e90003. doi: 10.1371/journal.pone.0090003. eCollection 2014. PLoS One. 2014. PMID: 24587180 Free PMC article.
-
Antiviral innate immunity and stress granule responses.Trends Immunol. 2014 Sep;35(9):420-8. doi: 10.1016/j.it.2014.07.006. Epub 2014 Aug 19. Trends Immunol. 2014. PMID: 25153707 Free PMC article. Review.
-
Control of the induction of type I interferon by Peste des petits ruminants virus.PLoS One. 2017 May 5;12(5):e0177300. doi: 10.1371/journal.pone.0177300. eCollection 2017. PLoS One. 2017. PMID: 28475628 Free PMC article.
-
Finding Gene Regulatory Networks in Psoriasis: Application of a Tree-Based Machine Learning Approach.Front Immunol. 2022 Jul 7;13:921408. doi: 10.3389/fimmu.2022.921408. eCollection 2022. Front Immunol. 2022. PMID: 35874668 Free PMC article.
References
-
- Griffin DE. 2007. Measles virus, p 1551–1585 In Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE. (ed), Fields virology, 5th ed. Lippincott/The Williams & Wilkins Co, Philadelphia, PA
-
- Cattaneo R, Kaelin K, Baczko K, Billeter MA. 1989. Measles virus editing provides an additional cysteine-rich protein. Cell 56:759–764 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

