Viral-host interactions that control HIV-1 transcriptional elongation
- PMID: 23795863
- PMCID: PMC4310557
- DOI: 10.1021/cr400120z
Viral-host interactions that control HIV-1 transcriptional elongation
Abstract
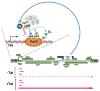
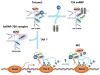
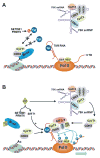
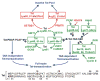
Regulation of the pause and elongation by RNA polymerase (Pol) II is used widely by metazoans to attain the pattern of gene expression that is essential for optimal cell growth/renewal, differentiation and stress response. Currently, much of what we know about Pol II elongation control comes from pioneering studies of the HIV-1-encoded Tat protein and its host cellular co-factors. The interaction between the two fuels a powerful feedback circuit that activates HIV transcription and prevents the virus from entering latency. One of the key Tat cofactors is the human positive transcription elongation factor b (P-TEFb), which exists in a family of complexes with distinct functions during Tat transactivation. This article reviews recent progresses in HIV transcription research with an emphasis on the intricate control of the various P-TEFb complexes, structural and functional insights into their interactions with Tat, the multifaceted roles of post-translational modifications of Tat and epigenetic control of HIV chromatin in modulating Tat activity and HIV latency. The knowledge from these studies will not only help design better strategies to fight HIV infection and transcriptional latency, but also advance the overall understanding of the mechanism controlling transcriptional elongation in general.
Figures


Similar articles
-
HIV-1 Tat assembles a multifunctional transcription elongation complex and stably associates with the 7SK snRNP.Mol Cell. 2010 May 14;38(3):439-51. doi: 10.1016/j.molcel.2010.04.012. Mol Cell. 2010. PMID: 20471949 Free PMC article.
-
Kick-sTARting HIV-1 transcription elongation by 7SK snRNP deporTATion.Nat Struct Mol Biol. 2010 Aug;17(8):928-30. doi: 10.1038/nsmb0810-928. Nat Struct Mol Biol. 2010. PMID: 20683478
-
Gene target specificity of the Super Elongation Complex (SEC) family: how HIV-1 Tat employs selected SEC members to activate viral transcription.Nucleic Acids Res. 2015 Jul 13;43(12):5868-79. doi: 10.1093/nar/gkv541. Epub 2015 May 24. Nucleic Acids Res. 2015. PMID: 26007649 Free PMC article.
-
New insights into the control of HIV-1 transcription: when Tat meets the 7SK snRNP and super elongation complex (SEC).J Neuroimmune Pharmacol. 2011 Jun;6(2):260-8. doi: 10.1007/s11481-011-9267-6. Epub 2011 Mar 1. J Neuroimmune Pharmacol. 2011. PMID: 21360054 Free PMC article. Review.
-
The control of HIV transcription: keeping RNA polymerase II on track.Cell Host Microbe. 2011 Nov 17;10(5):426-35. doi: 10.1016/j.chom.2011.11.002. Cell Host Microbe. 2011. PMID: 22100159 Free PMC article. Review.
Cited by
-
Hacking the Cell: Network Intrusion and Exploitation by Adenovirus E1A.mBio. 2018 May 1;9(3):e00390-18. doi: 10.1128/mBio.00390-18. mBio. 2018. PMID: 29717008 Free PMC article. Review.
-
AFF1 is a ubiquitous P-TEFb partner to enable Tat extraction of P-TEFb from 7SK snRNP and formation of SECs for HIV transactivation.Proc Natl Acad Sci U S A. 2014 Jan 7;111(1):E15-24. doi: 10.1073/pnas.1318503111. Epub 2013 Dec 23. Proc Natl Acad Sci U S A. 2014. PMID: 24367103 Free PMC article.
-
LARP7 suppresses P-TEFb activity to inhibit breast cancer progression and metastasis.Elife. 2014 Jul 22;3:e02907. doi: 10.7554/eLife.02907. Elife. 2014. PMID: 25053741 Free PMC article.
-
Insights into HIV-1 proviral transcription from integrative structure and dynamics of the Tat:AFF4:P-TEFb:TAR complex.Elife. 2016 Oct 12;5:e15910. doi: 10.7554/eLife.15910. Elife. 2016. PMID: 27731797 Free PMC article.
-
The Old and New Testaments of gene regulation. Evolution of multi-subunit RNA polymerases and co-evolution of eukaryote complexity with the RNAP II CTD.Transcription. 2014;5(3):e28674. doi: 10.4161/trns.28674. Transcription. 2014. PMID: 25764332 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

