The Us2 gene product of herpes simplex virus 2 is a membrane-associated ubiquitin-interacting protein
- PMID: 23785212
- PMCID: PMC3754132
- DOI: 10.1128/JVI.00994-13
The Us2 gene product of herpes simplex virus 2 is a membrane-associated ubiquitin-interacting protein
Abstract
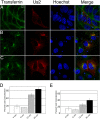
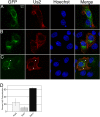
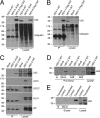
The Us2 gene encodes a tegument protein that is conserved in most members of the Alphaherpesvirinae. Previous studies on the pseudorabies virus (PRV) Us2 ortholog indicated that it is prenylated, associates with membranes, and spatially regulates the enzymatic activity of the MAP (mitogen-activated protein) kinase ERK (extracellular signal-related kinase) through direct binding and sequestration of ERK at the cytoplasmic face of the plasma membrane. Here we present an analysis of the herpes simplex virus 2 (HSV-2) Us2 ortholog and demonstrate that, like PRV Us2, HSV-2 Us2 is a virion component and that, unlike PRV Us2, it does not interact with ERK in yeast two-hybrid assays. HSV-2 Us2 lacks prenylation signals and other canonical membrane-targeting motifs yet is tightly associated with detergent-insoluble membranes and localizes predominantly to recycling endosomes. Experiments to identify cellular proteins that facilitate HSV-2 Us2 membrane association were inconclusive; however, these studies led to the identification of HSV-2 Us2 as a ubiquitin-interacting protein, providing new insight into the functions of HSV-2 Us2.
Figures
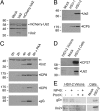
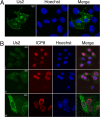
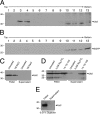

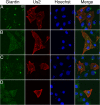

Similar articles
-
Pseudorabies virus tegument protein Us2 recruits the mitogen-activated protein kinase extracellular-regulated kinase (ERK) to membranes through interaction with the ERK common docking domain.J Virol. 2010 Sep;84(17):8398-408. doi: 10.1128/JVI.00794-10. Epub 2010 Jun 16. J Virol. 2010. PMID: 20554783 Free PMC article.
-
Localization of ERK/MAP kinase is regulated by the alphaherpesvirus tegument protein Us2.J Virol. 2006 Jul;80(14):7159-68. doi: 10.1128/JVI.00592-06. J Virol. 2006. PMID: 16809321 Free PMC article.
-
The US2 gene product of herpes simplex virus type 2 interacts with cytokeratin 18.Arch Virol. 2001;146(11):2201-9. doi: 10.1007/s007050170030. Arch Virol. 2001. PMID: 11765921
-
The Us2 Gene Product of Herpes Simplex Virus 2 modulates NF-κB activation by targeting TAK1.Sci Rep. 2017 Aug 21;7(1):8396. doi: 10.1038/s41598-017-08856-4. Sci Rep. 2017. PMID: 28827540 Free PMC article.
-
The pseudorabies virus Us2 protein, a virion tegument component, is prenylated in infected cells.J Virol. 2003 Nov;77(22):12285-98. doi: 10.1128/jvi.77.22.12285-12298.2003. J Virol. 2003. PMID: 14581565 Free PMC article.
Cited by
-
Pseudorabies virus tegument protein US2 antagonizes antiviral innate immunity by targeting cGAS-STING signaling pathway.Front Immunol. 2024 Jul 2;15:1403070. doi: 10.3389/fimmu.2024.1403070. eCollection 2024. Front Immunol. 2024. PMID: 39015575 Free PMC article.
-
Deletion of pseudorabies virus US2 gene enhances viral titers in a porcine cerebral cortex primary culture system.Virus Genes. 2018 Jun;54(3):406-413. doi: 10.1007/s11262-018-1552-5. Epub 2018 Mar 14. Virus Genes. 2018. PMID: 29541931
-
Phylogenetic and Genomic Characterization of Whole Genome Sequences of Ocular Herpes Simplex Virus Type 1 Isolates Identifies Possible Virulence Determinants in Humans.Invest Ophthalmol Vis Sci. 2023 Jul 3;64(10):16. doi: 10.1167/iovs.64.10.16. Invest Ophthalmol Vis Sci. 2023. PMID: 37450309 Free PMC article.
-
"Non-Essential" Proteins of HSV-1 with Essential Roles In Vivo: A Comprehensive Review.Viruses. 2020 Dec 23;13(1):17. doi: 10.3390/v13010017. Viruses. 2020. PMID: 33374862 Free PMC article. Review.
-
Epithelial Intermediate Filaments: Guardians against Microbial Infection?Cells. 2016 Jun 27;5(3):29. doi: 10.3390/cells5030029. Cells. 2016. PMID: 27355965 Free PMC article. Review.
References
-
- Ben-Porat T, Kaplan AS. 1985. Molecular biology of pseudorabies virus, p 105–173 In Roizman B. (ed), The herpesviruses. Plenum Press, New York, NY
-
- Roizman B, Knipe DM. 2001. Herpes simplex viruses and their replication, p 2399–2459 In Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE. (ed), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, PA
-
- Mettenleiter TC, Klupp BG, Granzow H. 2009. Herpesvirus assembly: an update. Virus Res. 143:222–234 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

