Transcription of the Escherichia coli fatty acid synthesis operon fabHDG is directly activated by FadR and inhibited by ppGpp
- PMID: 23772072
- PMCID: PMC3754556
- DOI: 10.1128/JB.00384-13
Transcription of the Escherichia coli fatty acid synthesis operon fabHDG is directly activated by FadR and inhibited by ppGpp
Abstract
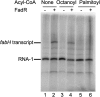
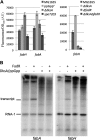
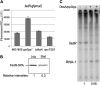
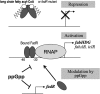
In Escherichia coli, FadR and FabR are transcriptional regulators that control the expression of fatty acid degradation and unsaturated fatty acid synthesis genes, depending on the availability of fatty acids. In this report, we focus on the dual transcriptional regulator FadR. In the absence of fatty acids, FadR represses the transcription of fad genes required for fatty acid degradation. However, FadR is also an activator, stimulating transcription of the products of the fabA and fabB genes responsible for unsaturated fatty acid synthesis. In this study, we show that FadR directly activates another fatty acid synthesis promoter, PfabH, which transcribes the fabHDG operon, indicating that FadR is a global regulator of both fatty acid degradation and fatty acid synthesis. We also demonstrate that ppGpp and its cofactor DksA, known primarily for their role in regulation of the synthesis of the translational machinery, directly inhibit transcription from the fabH promoter. ppGpp also inhibits the fadR promoter, thereby reducing transcription activation of fabH by FadR indirectly. Our study shows that both ppGpp and FadR have direct roles in the control of fatty acid promoters, linking expression in response to both translation activity and fatty acid availability.
Figures
Similar articles
-
Reassessment of the Genetic Regulation of Fatty Acid Synthesis in Escherichia coli: Global Positive Control by the Dual Functional Regulator FadR.J Bacteriol. 2015 Jun;197(11):1862-72. doi: 10.1128/JB.00064-15. Epub 2015 Mar 23. J Bacteriol. 2015. PMID: 25802297 Free PMC article.
-
Crosstalk of Escherichia coli FadR with global regulators in expression of fatty acid transport genes.PLoS One. 2012;7(9):e46275. doi: 10.1371/journal.pone.0046275. Epub 2012 Sep 28. PLoS One. 2012. PMID: 23029459 Free PMC article.
-
Complex binding of the FabR repressor of bacterial unsaturated fatty acid biosynthesis to its cognate promoters.Mol Microbiol. 2011 Apr;80(1):195-218. doi: 10.1111/j.1365-2958.2011.07564.x. Epub 2011 Feb 21. Mol Microbiol. 2011. PMID: 21276098 Free PMC article.
-
The Escherichia coli FadR transcription factor: Too much of a good thing?Mol Microbiol. 2021 Jun;115(6):1080-1085. doi: 10.1111/mmi.14663. Epub 2020 Dec 19. Mol Microbiol. 2021. PMID: 33283913 Free PMC article. Review.
-
Regulation of fatty acid metabolism in bacteria.Mol Microbiol. 2007 Nov;66(4):829-39. doi: 10.1111/j.1365-2958.2007.05947.x. Epub 2007 Oct 2. Mol Microbiol. 2007. PMID: 17919287 Review.
Cited by
-
The Classical, Yet Controversial, First Enzyme of Lipid Synthesis: Escherichia coli Acetyl-CoA Carboxylase.Microbiol Mol Biol Rev. 2021 Aug 18;85(3):e0003221. doi: 10.1128/MMBR.00032-21. Epub 2021 Jun 16. Microbiol Mol Biol Rev. 2021. PMID: 34132100 Free PMC article. Review.
-
Linking glucose metabolism to the stringent response through the PTS.Proc Natl Acad Sci U S A. 2018 Jul 17;115(29):7454-7455. doi: 10.1073/pnas.1809265115. Epub 2018 Jul 3. Proc Natl Acad Sci U S A. 2018. PMID: 29970419 Free PMC article. No abstract available.
-
A Novel Gene Contributing to the Initiation of Fatty Acid Biosynthesis in Escherichia coli.J Bacteriol. 2019 Sep 6;201(19):e00354-19. doi: 10.1128/JB.00354-19. Print 2019 Oct 1. J Bacteriol. 2019. PMID: 31331975 Free PMC article.
-
AzeR, a transcriptional regulator that responds to azelaic acid in Pseudomonas nitroreducens.Microbiology (Reading). 2020 Jan;166(1):73-84. doi: 10.1099/mic.0.000865. Microbiology (Reading). 2020. PMID: 31621557 Free PMC article.
-
RSH enzyme diversity for (p)ppGpp metabolism in Phaeodactylum tricornutum and other diatoms.Sci Rep. 2019 Nov 27;9(1):17682. doi: 10.1038/s41598-019-54207-w. Sci Rep. 2019. PMID: 31776430 Free PMC article.
References
-
- Zhang YM, Rock CO. 2008. Membrane lipid homeostasis in bacteria. Nat. Rev. Microbiol. 6:222–233 - PubMed
-
- DiRusso CC, Black PN, Weimar JD. 1999. Molecular inroads into the regulation and metabolism of fatty acids, lessons from bacteria. Prog. Lipid Res. 38:129–197 - PubMed
-
- Fujita Y, Matsuoka H, Hirooka K. 2007. Regulation of fatty acid metabolism in bacteria. Mol. Microbiol. 66:829–839 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

