α-Amino-3-hydroxy-5-methyl-4-isoxazole propionic acid (AMPA) and N-methyl-D-aspartate (NMDA) receptors adopt different subunit arrangements
- PMID: 23760273
- PMCID: PMC3724652
- DOI: 10.1074/jbc.M113.469205
α-Amino-3-hydroxy-5-methyl-4-isoxazole propionic acid (AMPA) and N-methyl-D-aspartate (NMDA) receptors adopt different subunit arrangements
Abstract
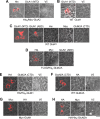
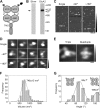
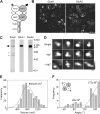
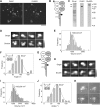
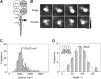
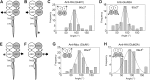
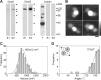
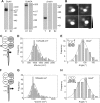
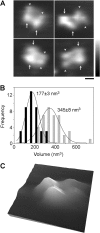
Ionotropic glutamate receptors are widely distributed in the central nervous system and play a major role in excitatory synaptic transmission. All three ionotropic glutamate subfamilies (i.e. AMPA-type, kainate-type, and NMDA-type) assemble as tetramers of four homologous subunits. There is good evidence that both heteromeric AMPA and kainate receptors have a 2:2 subunit stoichiometry and an alternating subunit arrangement. Recent studies based on presumed structural homology have indicated that NMDA receptors adopt the same arrangement. Here, we use atomic force microscopy imaging of receptor-antibody complexes to show that whereas the GluA1/GluA2 AMPA receptor assembles with an alternating (i.e. 1/2/1/2) subunit arrangement, the GluN1/GluN2A NMDA receptor adopts an adjacent (i.e. 1/1/2/2) arrangement. We conclude that the two types of ionotropic glutamate receptor are built in different ways from their constituent subunits. This surprising finding necessitates a reassessment of the assembly of these important receptors.
Keywords: Atomic Force Microscopy; Cell Surface Receptor; Glutamate Receptors, Ionotropic (AMPA, NMDA); Protein Complexes; Single-particle Analysis.
Figures
Similar articles
-
An alternating GluN1-2-1-2 subunit arrangement in mature NMDA receptors.PLoS One. 2012;7(4):e35134. doi: 10.1371/journal.pone.0035134. Epub 2012 Apr 6. PLoS One. 2012. PMID: 22493736 Free PMC article.
-
Localization of AMPA, kainate, and NMDA receptor mRNAs in the pigeon cerebellum.J Chem Neuroanat. 2019 Jul;98:71-79. doi: 10.1016/j.jchemneu.2019.04.004. Epub 2019 Apr 9. J Chem Neuroanat. 2019. PMID: 30978490
-
Nucleus-specific expression of ionotropic glutamate receptor subunit mRNAs and binding sites in primate thalamus.Brain Res Mol Brain Res. 2000 Jun 23;79(1-2):1-17. doi: 10.1016/s0169-328x(00)00072-3. Brain Res Mol Brain Res. 2000. PMID: 10925139
-
Structural dynamics in α-amino-3-hydroxy-5-methyl-4-isoxazole propionic acid receptor gating.Curr Opin Struct Biol. 2024 Aug;87:102833. doi: 10.1016/j.sbi.2024.102833. Epub 2024 May 10. Curr Opin Struct Biol. 2024. PMID: 38733862 Review.
-
The biochemistry, ultrastructure, and subunit assembly mechanism of AMPA receptors.Mol Neurobiol. 2010 Dec;42(3):161-84. doi: 10.1007/s12035-010-8149-x. Epub 2010 Nov 16. Mol Neurobiol. 2010. PMID: 21080238 Free PMC article. Review.
Cited by
-
Radial symmetry in a chimeric glutamate receptor pore.Nat Commun. 2014;5:3349. doi: 10.1038/ncomms4349. Nat Commun. 2014. PMID: 24561802 Free PMC article.
-
Aggregation Limits Surface Expression of Homomeric GluA3 Receptors.J Biol Chem. 2016 Apr 15;291(16):8784-94. doi: 10.1074/jbc.M115.689125. Epub 2016 Feb 24. J Biol Chem. 2016. PMID: 26912664 Free PMC article.
-
Role of actin cytoskeleton in the organization and function of ionotropic glutamate receptors.Curr Res Struct Biol. 2021 Oct 14;3:277-289. doi: 10.1016/j.crstbi.2021.10.001. eCollection 2021. Curr Res Struct Biol. 2021. PMID: 34766008 Free PMC article. Review.
-
Glutamate receptor pores.J Physiol. 2015 Jan 1;593(1):49-59. doi: 10.1113/jphysiol.2014.272724. Epub 2014 May 6. J Physiol. 2015. PMID: 25556787 Free PMC article. Review.
-
Inhibitory interactions between phosphorylation sites in the C terminus of α-Amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid-type glutamate receptor GluA1 subunits.J Biol Chem. 2014 May 23;289(21):14600-11. doi: 10.1074/jbc.M114.553537. Epub 2014 Apr 4. J Biol Chem. 2014. PMID: 24706758 Free PMC article.
References
-
- Mansour M., Nagarajan N., Nehring R. B., Clements J. D., Rosenmund C. (2001) Heteromeric AMPA receptors assemble with a preferred subunit stoichiometry and spatial arrangement. Neuron 32, 841–853 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

