Quantifying the topography of the intrinsic energy landscape of flexible biomolecular recognition
- PMID: 23754431
- PMCID: PMC3696809
- DOI: 10.1073/pnas.1220699110
Quantifying the topography of the intrinsic energy landscape of flexible biomolecular recognition
Abstract
Biomolecular functions are determined by their interactions with other molecules. Biomolecular recognition is often flexible and associated with large conformational changes involving both binding and folding. However, the global and physical understanding for the process is still challenging. Here, we quantified the intrinsic energy landscapes of flexible biomolecular recognition in terms of binding-folding dynamics for 15 homodimers by exploring the underlying density of states, using a structure-based model both with and without considering energetic roughness. By quantifying three individual effective intrinsic energy landscapes (one for interfacial binding, two for monomeric folding), the association mechanisms for flexible recognition of 15 homodimers can be classified into two-state cooperative "coupled binding-folding" and three-state noncooperative "folding prior to binding" scenarios. We found that the association mechanism of flexible biomolecular recognition relies on the interplay between the underlying effective intrinsic binding and folding energy landscapes. By quantifying the whole global intrinsic binding-folding energy landscapes, we found strong correlations between the landscape topography measure Λ (dimensionless ratio of energy gap versus roughness modulated by the configurational entropy) and the ratio of the thermodynamic stable temperature versus trapping temperature, as well as between Λ and binding kinetics. Therefore, the global energy landscape topography determines the binding-folding thermodynamics and kinetics, crucial for the feasibility and efficiency of realizing biomolecular function. We also found "U-shape" temperature-dependent kinetic behavior and a dynamical cross-over temperature for dividing exponential and nonexponential kinetics for two-state homodimers. Our study provides a unique way to bridge the gap between theory and experiments.
Keywords: energy landscape theory; flexible binding-folding; intrinsically disordered proteins.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
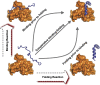
 and
and  are the energy of interfacial binding and monomeric folding interaction. Logarithm of DOS is plotted as a function of Q for (C) Arc repressor and (D) Lambda Cro repressor.
are the energy of interfacial binding and monomeric folding interaction. Logarithm of DOS is plotted as a function of Q for (C) Arc repressor and (D) Lambda Cro repressor.  and
and  are the fraction of native binding and folding contacts. Because the homodimers are formed by two identical chains, the folding properties of the two monomers are expected to be same. Here, we use quantities of one of the two monomers to represent the corresponding monomeric folding quantities. Energy is plotted as a function of
are the fraction of native binding and folding contacts. Because the homodimers are formed by two identical chains, the folding properties of the two monomers are expected to be same. Here, we use quantities of one of the two monomers to represent the corresponding monomeric folding quantities. Energy is plotted as a function of  and
and  for (E) Arc repressor and (F) Lambda Cro repressor. White region is not probed by the binding–folding simulations. Energy is in the reduced unit (73).
for (E) Arc repressor and (F) Lambda Cro repressor. White region is not probed by the binding–folding simulations. Energy is in the reduced unit (73).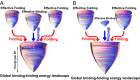
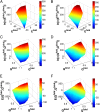
 , and a monotonic decreasing function of the fraction of native contacts Q,and
, and a monotonic decreasing function of the fraction of native contacts Q,and  is a scaling constant for each funnel to achieve a better visualization. The other semi-axis of the ellipse is therefore given by
is a scaling constant for each funnel to achieve a better visualization. The other semi-axis of the ellipse is therefore given by  . The unbound states are located at the top of the funnels with the largest energy, entropy, and smallest Q, and the bottom of the funnel corresponds to the native binding state with
. The unbound states are located at the top of the funnels with the largest energy, entropy, and smallest Q, and the bottom of the funnel corresponds to the native binding state with  and
and  . The shape of the funnels is therefore a representation of the relationship between energy E, entropy S, and structural similarity to native structure Q. The energy and entropy are normalized by the sizes of the homodimers for a better visualization.
. The shape of the funnels is therefore a representation of the relationship between energy E, entropy S, and structural similarity to native structure Q. The energy and entropy are normalized by the sizes of the homodimers for a better visualization.
 and monomeric folding
and monomeric folding  . The PDB ID codes of the 15 homodimers are shown.
. The PDB ID codes of the 15 homodimers are shown.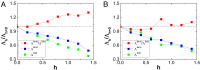
 versus
versus  , which corresponds to the pure structure-based model without nonnative interactions, for (A) Arc repressor and (B) Lambda Cro repressor. Red points represent the ratio of
, which corresponds to the pure structure-based model without nonnative interactions, for (A) Arc repressor and (B) Lambda Cro repressor. Red points represent the ratio of  , blue points represent the ratio of
, blue points represent the ratio of  , and green points represent the ratio of
, and green points represent the ratio of  as a function of strength of nonnative interactions. The parameter h modulates the strength of nonnative interactions.
as a function of strength of nonnative interactions. The parameter h modulates the strength of nonnative interactions.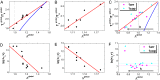
 and thermodynamics as well as kinetics of global binding–folding. The binding transition against glassy trapping temperature
and thermodynamics as well as kinetics of global binding–folding. The binding transition against glassy trapping temperature  correlated with the whole global landscape topography
correlated with the whole global landscape topography  for (A) 10 different-sized homodimers with correlation 0.94, (B) eight similar-sized homodimers with correlation 0.84, and (C) two-state homodimer Arc repressor and three-state homodimer Lambda Cro repressor with varying energetic roughness, showing a correlation of 0.88. The blue lines in A, B, and C are the analytical mean field theoretical prediction of the relationship between
for (A) 10 different-sized homodimers with correlation 0.94, (B) eight similar-sized homodimers with correlation 0.84, and (C) two-state homodimer Arc repressor and three-state homodimer Lambda Cro repressor with varying energetic roughness, showing a correlation of 0.88. The blue lines in A, B, and C are the analytical mean field theoretical prediction of the relationship between  and
and  . The logarithm of global binding–folding time correlated with the whole global landscape topography
. The logarithm of global binding–folding time correlated with the whole global landscape topography  for (D) 10 different-sized homodimers with correction –0.78, (E) eight similar-sized homodimers with correlation –0.91, and (F) two-state homodimer Arc repressor and three-state homodimer Lambda Cro repressor with varying energetic roughness. The red lines in A–E are the linear fits. Because all of the quantities are found to be size scaled (
for (D) 10 different-sized homodimers with correction –0.78, (E) eight similar-sized homodimers with correlation –0.91, and (F) two-state homodimer Arc repressor and three-state homodimer Lambda Cro repressor with varying energetic roughness. The red lines in A–E are the linear fits. Because all of the quantities are found to be size scaled ( and thermodynamics, as well as kinetics. The parameter h modulates the strength of nonnative interactions and τ is in the unit of simulation time.
and thermodynamics, as well as kinetics. The parameter h modulates the strength of nonnative interactions and τ is in the unit of simulation time.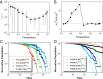
 as a function of temperature. The error bar represents the SD of the mean value. (B) The second-order moment ratio of the binding–folding time
as a function of temperature. The error bar represents the SD of the mean value. (B) The second-order moment ratio of the binding–folding time  versus temperature. (C) The evolution of nonnative population at different temperatures above the activated dynamical cross-over temperature
versus temperature. (C) The evolution of nonnative population at different temperatures above the activated dynamical cross-over temperature  . (D) The evolution of nonnative population at different temperatures below the activated dynamical cross-over temperature
. (D) The evolution of nonnative population at different temperatures below the activated dynamical cross-over temperature  . The fitting lines in C and D are single exponential. Temperature is in the unit of energy by multiplying Boltzmann constant k, and τ is in the unit of simulation time.
. The fitting lines in C and D are single exponential. Temperature is in the unit of energy by multiplying Boltzmann constant k, and τ is in the unit of simulation time.Similar articles
-
Quantifying the Intrinsic Conformation Energy Landscape Topography of Proteins with Large-Scale Open-Closed Transition.ACS Cent Sci. 2018 Aug 22;4(8):1015-1022. doi: 10.1021/acscentsci.8b00274. Epub 2018 Aug 1. ACS Cent Sci. 2018. PMID: 30159398 Free PMC article.
-
Specificity and affinity quantification of flexible recognition from underlying energy landscape topography.PLoS Comput Biol. 2014 Aug 21;10(8):e1003782. doi: 10.1371/journal.pcbi.1003782. eCollection 2014 Aug. PLoS Comput Biol. 2014. PMID: 25144525 Free PMC article.
-
Topography of funneled landscapes determines the thermodynamics and kinetics of protein folding.Proc Natl Acad Sci U S A. 2012 Sep 25;109(39):15763-8. doi: 10.1073/pnas.1212842109. Epub 2012 Sep 10. Proc Natl Acad Sci U S A. 2012. PMID: 23019359 Free PMC article.
-
Physics of biomolecular recognition and conformational dynamics.Rep Prog Phys. 2021 Dec 8;84(12). doi: 10.1088/1361-6633/ac3800. Rep Prog Phys. 2021. PMID: 34753115 Review.
-
From protein folding to protein function and biomolecular binding by energy landscape theory.Curr Opin Pharmacol. 2010 Dec;10(6):709-14. doi: 10.1016/j.coph.2010.09.012. Epub 2010 Oct 14. Curr Opin Pharmacol. 2010. PMID: 20951644 Review.
Cited by
-
Structure, stability and specificity of the binding of ssDNA and ssRNA with proteins.PLoS Comput Biol. 2019 Apr 1;15(4):e1006768. doi: 10.1371/journal.pcbi.1006768. eCollection 2019 Apr. PLoS Comput Biol. 2019. PMID: 30933978 Free PMC article.
-
Protein recognition and selection through conformational and mutually induced fit.Proc Natl Acad Sci U S A. 2013 Dec 17;110(51):20545-50. doi: 10.1073/pnas.1312788110. Epub 2013 Dec 2. Proc Natl Acad Sci U S A. 2013. PMID: 24297894 Free PMC article.
-
Inferring potential landscapes from noisy trajectories of particles within an optical feedback trap.iScience. 2022 Jul 19;25(9):104731. doi: 10.1016/j.isci.2022.104731. eCollection 2022 Sep 16. iScience. 2022. PMID: 36034218 Free PMC article.
-
Conformational state switching and pathways of chromosome dynamics in cell cycle.Appl Phys Rev. 2020 Sep;7(3):031403. doi: 10.1063/5.0007316. Appl Phys Rev. 2020. PMID: 32884608 Free PMC article.
-
Electrostatic forces govern the binding mechanism of intrinsically disordered histone chaperones.PLoS One. 2017 May 26;12(5):e0178405. doi: 10.1371/journal.pone.0178405. eCollection 2017. PLoS One. 2017. PMID: 28552960 Free PMC article.
References
-
- Fischer E. Einfluss der configuration auf die wirkung der enzyme. Ber Dtsch Chem Ges. 1894;27(3):2984–2993.
-
- Bosshard HR. Molecular recognition by induced fit: How fit is the concept? News Physiol Sci. 2001;16(4):171–173. - PubMed
-
- Wright PE, Dyson HJ. Intrinsically unstructured proteins: Re-assessing the protein structure-function paradigm. J Mol Biol. 1999;293(2):321–331. - PubMed
-
- Dunker AK, et al. Intrinsically disordered protein. J Mol Graph Model. 2001;19(1):26–59. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

