The role of recombination in the origin and evolution of Alu subfamilies
- PMID: 23750218
- PMCID: PMC3672193
- DOI: 10.1371/journal.pone.0064884
The role of recombination in the origin and evolution of Alu subfamilies
Abstract
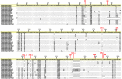
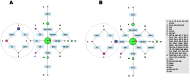
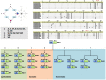
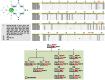
Alus are the most abundant and successful short interspersed nuclear elements found in primate genomes. In humans, they represent about 10% of the genome, although few are retrotransposition-competent and are clustered into subfamilies according to the source gene from which they evolved. Recombination between them can lead to genomic rearrangements of clinical and evolutionary significance. In this study, we have addressed the role of recombination in the origin of chimeric Alu source genes by the analysis of all known consensus sequences of human Alus. From the allelic diversity of Alu consensus sequences, validated in extant elements resulting from whole genome searches, distinct events of recombination were detected in the origin of particular subfamilies of AluS and AluY source genes. These results demonstrate that at least two subfamilies are likely to have emerged from ectopic Alu-Alu recombination, which stimulates further research regarding the potential of chimeric active Alus to punctuate the genome.
Conflict of interest statement
Figures

Similar articles
-
The biased distribution of Alus in human isochores might be driven by recombination.J Mol Evol. 2005 Mar;60(3):365-77. doi: 10.1007/s00239-004-0197-2. J Mol Evol. 2005. PMID: 15871047
-
Identification of human-specific AluS elements through comparative genomics.Gene. 2015 Jan 25;555(2):208-16. doi: 10.1016/j.gene.2014.11.005. Epub 2014 Nov 7. Gene. 2015. PMID: 25447892
-
Alu repeats and human genomic diversity.Nat Rev Genet. 2002 May;3(5):370-9. doi: 10.1038/nrg798. Nat Rev Genet. 2002. PMID: 11988762 Review.
-
Evolution of Alu Subfamily Structure in the Saimiri Lineage of New World Monkeys.Genome Biol Evol. 2017 Sep 1;9(9):2365-2376. doi: 10.1093/gbe/evx172. Genome Biol Evol. 2017. PMID: 28957461 Free PMC article.
-
Alu elements and the human genome.Genetica. 2000;108(1):57-72. doi: 10.1023/a:1004099605261. Genetica. 2000. PMID: 11145422 Review.
Cited by
-
Major influence of repetitive elements on disease-associated copy number variants (CNVs).Hum Genomics. 2016 Sep 23;10(1):30. doi: 10.1186/s40246-016-0088-9. Hum Genomics. 2016. PMID: 27663310 Free PMC article. Review.
-
Transposable elements that have recently been mobile in the human genome.BMC Genomics. 2021 Nov 3;22(1):789. doi: 10.1186/s12864-021-08085-0. BMC Genomics. 2021. PMID: 34732136 Free PMC article.
-
Alu RNA fold links splicing with signal recognition particle proteins.Nucleic Acids Res. 2023 Aug 25;51(15):8199-8216. doi: 10.1093/nar/gkad500. Nucleic Acids Res. 2023. PMID: 37309897 Free PMC article.
-
DNA word analysis based on the distribution of the distances between symmetric words.Sci Rep. 2017 Apr 7;7(1):728. doi: 10.1038/s41598-017-00646-2. Sci Rep. 2017. PMID: 28389642 Free PMC article.
-
Sequence Analysis and Characterization of Active Human Alu Subfamilies Based on the 1000 Genomes Pilot Project.Genome Biol Evol. 2015 Aug 29;7(9):2608-22. doi: 10.1093/gbe/evv167. Genome Biol Evol. 2015. PMID: 26319576 Free PMC article.
References
-
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, et al. (2001) Initial sequencing and analysis of the human genome. Nature 409: 860–921. - PubMed
-
- Ullu E, Tschudi C (1984) Alu sequences are processed 7SL RNA genes. Nature 312: 171–172. - PubMed
-
- Rogers JH (1985) The origin and evolution of retroposons. International Review of Cytology-a Survey of Cell Biology 93: 187–279. - PubMed
-
- Weiner AM, Deininger PL, Efstratiadis A (1986) nonviral retroposons - genes, pseudogenes, and transposable elements generated by the reverse flow of genetic information. Annual Review of Biochemistry 55: 631–661. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

