SOMA: a single oligonucleotide mutagenesis and cloning approach
- PMID: 23750217
- PMCID: PMC3672168
- DOI: 10.1371/journal.pone.0064870
SOMA: a single oligonucleotide mutagenesis and cloning approach
Abstract
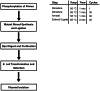
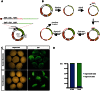
Modern biology research requires simple techniques for efficient and restriction site-independent modification of genetic material. Classical cloning and mutagenesis strategies are limited by their dependency on restriction sites and the use of complementary primer pairs. Here, we describe the Single Oligonucleotide Mutagenesis and Cloning Approach (SOMA) that is independent of restriction sites and only requires a single mutagenic oligonucleotide to modify a plasmid. We demonstrate the broad application spectrum of SOMA with three examples. First, we present a novel plasmid that in a standardized and rapid fashion can be used as a template for SOMA to generate GFP-reporters. We successfully use such a reporter to assess the in vivo knock-down quality of morpholinos in Xenopus laevis embryos. In a second example, we show how to use a SOMA-based protocol for restriction-site independent cloning to generate chimeric proteins by domain swapping between the two human hRMD5a and hRMD5b isoforms. Last, we show that SOMA simplifies the generation of randomized single-site mutagenized gene libraries. As an example we random-mutagenize a single codon affecting the catalytic activity of the yeast Ssy5 endoprotease and identify a spectrum of tolerated and non-tolerated substitutions. Thus, SOMA represents a highly efficient alternative to classical cloning and mutagenesis strategies.
Conflict of interest statement
Figures


Similar articles
-
Use of oligonucleotides and nick translation for site-directed mutagenesis in plasmids.Nucleic Acids Res. 1992 Feb 25;20(4):922. doi: 10.1093/nar/20.4.922. Nucleic Acids Res. 1992. PMID: 1542589 Free PMC article. No abstract available.
-
Method for cloning single-stranded oligonucleotides in a plasmid vector.Biotechniques. 1989 Apr;7(4):356-9. Biotechniques. 1989. PMID: 2698200
-
Ssy5 is a signaling serine protease that exhibits atypical biogenesis and marked S1 specificity.J Biol Chem. 2018 Jun 1;293(22):8362-8378. doi: 10.1074/jbc.RA118.002457. Epub 2018 Apr 16. J Biol Chem. 2018. PMID: 29661936 Free PMC article.
-
A QuikChange-like method to realize efficient blunt-ended DNA directional cloning and site-directed mutagenesis simultaneously.Biochem Biophys Res Commun. 2010 Jun 25;397(2):136-9. doi: 10.1016/j.bbrc.2010.05.042. Epub 2010 May 13. Biochem Biophys Res Commun. 2010. PMID: 20471367
-
Polishing the craft of genetic diversity creation in directed evolution.Biotechnol Adv. 2013 Dec;31(8):1707-21. doi: 10.1016/j.biotechadv.2013.08.021. Epub 2013 Sep 6. Biotechnol Adv. 2013. PMID: 24012599 Review.
Cited by
-
Cilia-localized GID/CTLH ubiquitin ligase complex regulates protein homeostasis of sonic hedgehog signaling components.J Cell Sci. 2022 May 1;135(9):jcs259209. doi: 10.1242/jcs.259209. Epub 2022 May 11. J Cell Sci. 2022. PMID: 35543155 Free PMC article.
-
Cytoplasmic protein misfolding titrates Hsp70 to activate nuclear Hsf1.Elife. 2019 Sep 25;8:e47791. doi: 10.7554/eLife.47791. Elife. 2019. PMID: 31552827 Free PMC article.
-
Interplay of TRIM2 E3 Ubiquitin Ligase and ALIX/ESCRT Complex: Control of Developmental Plasticity During Early Neurogenesis.Cells. 2020 Jul 20;9(7):1734. doi: 10.3390/cells9071734. Cells. 2020. PMID: 32698497 Free PMC article.
-
Regulation of MRTF-A by JMY via a nucleation-independent mechanism.Cell Commun Signal. 2018 Nov 21;16(1):86. doi: 10.1186/s12964-018-0299-x. Cell Commun Signal. 2018. PMID: 30463620 Free PMC article.
-
Post-transcriptional regulation of MRTF-A by miRNAs during myogenic differentiation of myoblasts.Nucleic Acids Res. 2020 Sep 18;48(16):8927-8942. doi: 10.1093/nar/gkaa596. Nucleic Acids Res. 2020. PMID: 32692361 Free PMC article.
References
-
- Pleiss J (2011) Protein design in metabolic engineering and synthetic biology. Curr Opin Biotechnol 22: 611–617. - PubMed
-
- Antikainen NM, Martin SF (2005) Altering protein specificity: techniques and applications. Bioorg Med Chem 13: 2701–2716. - PubMed
-
- Zawaira A, Pooran A, Barichievy S, Chopera D (2012) A discussion of molecular biology methods for protein engineering. Mol Biotechnol 51: 67–102. - PubMed
-
- Braman J, Papworth C, Greener A (1996) Site-directed mutagenesis using double-stranded plasmid DNA templates. Methods Mol Biol 57: 31–44. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

