Mapping the LINE1 ORF1 protein interactome reveals associated inhibitors of human retrotransposition
- PMID: 23749060
- PMCID: PMC3753637
- DOI: 10.1093/nar/gkt512
Mapping the LINE1 ORF1 protein interactome reveals associated inhibitors of human retrotransposition
Erratum in
-
Correction to 'Mapping the LINE1 ORF1 protein interactome reveals associated inhibitors of human retrotransposition'.Nucleic Acids Res. 2024 Feb 28;52(4):2091. doi: 10.1093/nar/gkae043. Nucleic Acids Res. 2024. PMID: 38261987 Free PMC article. No abstract available.
Abstract
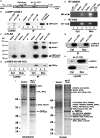
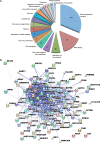
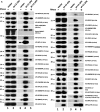
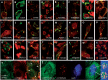
LINE1s occupy 17% of the human genome and are its only active autonomous mobile DNA. L1s are also responsible for genomic insertion of processed pseudogenes and >1 million non-autonomous retrotransposons (Alus and SVAs). These elements have significant effects on gene organization and expression. Despite the importance of retrotransposons for genome evolution, much about their biology remains unknown, including cellular factors involved in the complex processes of retrotransposition and forming and transporting L1 ribonucleoprotein particles. By co-immunoprecipitation of tagged L1 constructs and mass spectrometry, we identified proteins associated with the L1 ORF1 protein and its ribonucleoprotein. These include RNA transport proteins, gene expression regulators, post-translational modifiers, helicases and splicing factors. Many cellular proteins co-localize with L1 ORF1 protein in cytoplasmic granules. We also assayed the effects of these proteins on cell culture retrotransposition and found strong inhibiting proteins, including some that control HIV and other retroviruses. These data suggest candidate cofactors that interact with the L1 to modulate its activity and increase our understanding of the means by which the cell coexists with these genomic 'parasites'.
Figures


Similar articles
-
The Broad-Spectrum Antiviral Protein ZAP Restricts Human Retrotransposition.PLoS Genet. 2015 May 22;11(5):e1005252. doi: 10.1371/journal.pgen.1005252. eCollection 2015 May. PLoS Genet. 2015. PMID: 26001115 Free PMC article.
-
MOV10 RNA helicase is a potent inhibitor of retrotransposition in cells.PLoS Genet. 2012;8(10):e1002941. doi: 10.1371/journal.pgen.1002941. Epub 2012 Oct 18. PLoS Genet. 2012. PMID: 23093941 Free PMC article.
-
Discrete subcellular partitioning of human retrotransposon RNAs despite a common mechanism of genome insertion.Hum Mol Genet. 2010 May 1;19(9):1712-25. doi: 10.1093/hmg/ddq048. Epub 2010 Feb 10. Hum Mol Genet. 2010. PMID: 20147320 Free PMC article.
-
L1 elements, processed pseudogenes and retrogenes in mammalian genomes.IUBMB Life. 2006 Dec;58(12):677-85. doi: 10.1080/15216540601034856. IUBMB Life. 2006. PMID: 17424906 Review.
-
Human L1 retrotransposition: insights and peculiarities learned from a cultured cell retrotransposition assay.Genetica. 1999;107(1-3):39-51. Genetica. 1999. PMID: 10952196 Review.
Cited by
-
SAMHD1 Inhibits LINE-1 Retrotransposition by Promoting Stress Granule Formation.PLoS Genet. 2015 Jul 2;11(7):e1005367. doi: 10.1371/journal.pgen.1005367. eCollection 2015 Jul. PLoS Genet. 2015. PMID: 26134849 Free PMC article.
-
APOBEC3DE Inhibits LINE-1 Retrotransposition by Interacting with ORF1p and Influencing LINE Reverse Transcriptase Activity.PLoS One. 2016 Jul 18;11(7):e0157220. doi: 10.1371/journal.pone.0157220. eCollection 2016. PLoS One. 2016. PMID: 27428332 Free PMC article.
-
Reactivity of IgG With the p40 Protein Encoded by the Long Interspersed Nuclear Element 1 Retroelement: Comment on the Article by Carter et al.Arthritis Rheumatol. 2020 Feb;72(2):374-376. doi: 10.1002/art.41102. Epub 2019 Dec 27. Arthritis Rheumatol. 2020. PMID: 31513361 Free PMC article. No abstract available.
-
In vitro screening for compounds that enhance human L1 mobilization.PLoS One. 2013 Sep 11;8(9):e74629. doi: 10.1371/journal.pone.0074629. eCollection 2013. PLoS One. 2013. PMID: 24040300 Free PMC article.
-
Proteome Profile of Endogenous Retrotransposon-Associated Complexes in Human Embryonic Stem Cells.Proteomics. 2019 Aug;19(15):e1900169. doi: 10.1002/pmic.201900169. Epub 2019 Jul 18. Proteomics. 2019. PMID: 31219246 Free PMC article.
References
-
- Luan DD, Korman MH, Jakubczak JL, Eickbush TH. Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: a mechanism for non-LTR retrotransposition. Cell. 1993;72:595–605. - PubMed
-
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. - PubMed
-
- Wang H, Xing J, Grover D, Hedges DJ, Han K, Walker JA, Batzer MA. SVA elements: a hominid-specific retroposon family. J. Mol. Biol. 2005;354:994–1007. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

