RUNX3 gene promoter demethylation by 5-Aza-CdR induces apoptosis in breast cancer MCF-7 cell line
- PMID: 23723708
- PMCID: PMC3665559
- DOI: 10.2147/OTT.S43744
RUNX3 gene promoter demethylation by 5-Aza-CdR induces apoptosis in breast cancer MCF-7 cell line
Abstract
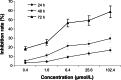
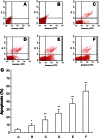
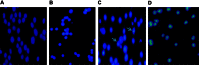
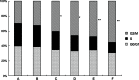
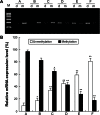
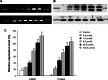
Runt-related transcription factor 3 (RUNX3) is a tumor suppressor gene, its inactivation due to hypermethylation related to carcinogenesis. The aim of this study was to investigate the effects of 5-aza-2'-deoxycytidine (5-Aza-CdR) on cell proliferation and apoptosis by demethylation of the promoter region and restoring the expression of RUNX3 in the breast cancer MCF-7 cell line. MCF-7 cells were cultured with different concentrations (0.4-102.4 μmol/L) of 5-Aza-CdR in vitro. MTT assay was used to determine the proliferation of MCF-7 cells. Flow cytometry and Hoechst staining were used for analyzing cell apoptosis. The methylation status and expression of RUNX3 in mRNA and protein levels were measured by methylation-specific polymerase chain reaction (PCR [MSP]), reverse transcription (RT)-PCR, and Western blot. It was shown that the RUNX3 gene downregulated and hypermethylated in MCF-7 cells. 5-Aza-CdR induced demethylation, upregulated the expression of RUNX3 on both mRNA and protein levels in cancer cells, and induced growth suppression and apoptosis in vitro in a dose- and time-dependent manner. The results demonstrate that RUNX3 downregulation in breast cancer is frequently due to hypermethylation, and that 5-Aza-CdR can inhibit cell proliferation and induce apoptosis by eliminating the methylation status of RUNX3 promoter and restoring its expression.
Keywords: RUNX3; apoptosis; breast cancer; methylation.
Figures
Similar articles
-
[Effect of 5-aza-2'-deoxycytidine on growth and methylation of RUNX3 gene in human pancreatic cancer cell line MiaPaca2].Zhonghua Zhong Liu Za Zhi. 2013 Jan;35(1):17-21. doi: 10.3760/cma.j.issn.0253-3766.2013.01.004. Zhonghua Zhong Liu Za Zhi. 2013. PMID: 23648294 Chinese.
-
[5-aza-2'-deoxycytidine induces changes of histone H3-lysine 9 methylation in bladder tumor cells].Zhonghua Yi Xue Za Zhi. 2008 Aug 19;88(32):2295-8. Zhonghua Yi Xue Za Zhi. 2008. PMID: 19087683 Chinese.
-
Effect of 5-aza-2'-deoxycytidine on cell proliferation of non- small cell lung cancer cell line A549 cells and expression of the TFPI-2 gene.Asian Pac J Cancer Prev. 2013;14(7):4421-6. doi: 10.7314/apjcp.2013.14.7.4421. Asian Pac J Cancer Prev. 2013. PMID: 23992014
-
Effects of 5-Aza-CdR on the proliferation of human breast cancer cell line MCF-7 and on the expression of Apaf-1 gene.J Huazhong Univ Sci Technolog Med Sci. 2009 Aug;29(4):498-502. doi: 10.1007/s11596-009-0421-9. Epub 2009 Aug 7. J Huazhong Univ Sci Technolog Med Sci. 2009. PMID: 19662370
-
RUNX3 is involved in caspase-3-dependent apoptosis induced by a combination of 5-aza-CdR and TSA in leukaemia cell lines.J Cancer Res Clin Oncol. 2012 Mar;138(3):439-49. doi: 10.1007/s00432-011-1113-y. Epub 2011 Dec 18. J Cancer Res Clin Oncol. 2012. PMID: 22179198
Cited by
-
Truncated forms of RUNX3 Unlike Full Length Protein Alter Cell Proliferation in a TGF-β Context Dependent Manner.Iran J Pharm Res. 2017 Summer;16(3):1194-1203. Iran J Pharm Res. 2017. PMID: 29201108 Free PMC article.
-
Regulation of RUNX3 Expression by DNA Methylation in Prostate Cancer.Cancer Manag Res. 2020 Jul 27;12:6411-6420. doi: 10.2147/CMAR.S249066. eCollection 2020. Cancer Manag Res. 2020. Retraction in: Cancer Manag Res. 2021 Dec 30;13:9421-9422. doi: 10.2147/CMAR.S355498 PMID: 32801881 Free PMC article. Retracted.
-
Baicalein increases the expression and reciprocal interplay of RUNX3 and FOXO3a through crosstalk of AMPKα and MEK/ERK1/2 signaling pathways in human non-small cell lung cancer cells.J Exp Clin Cancer Res. 2015 May 7;34(1):41. doi: 10.1186/s13046-015-0160-7. J Exp Clin Cancer Res. 2015. PMID: 25948105 Free PMC article.
-
Low concentrations of 5-aza-2'-deoxycytidine induce breast cancer stem cell differentiation by triggering tumor suppressor gene expression.Onco Targets Ther. 2015 Dec 23;9:49-59. doi: 10.2147/OTT.S96291. eCollection 2016. Onco Targets Ther. 2015. PMID: 26730203 Free PMC article.
-
Methylation status of the FHIT gene in the transformed human mesenchymal F6 stem cell line.Oncol Lett. 2015 Jun;9(6):2661-2666. doi: 10.3892/ol.2015.3092. Epub 2015 Apr 1. Oncol Lett. 2015. PMID: 26137124 Free PMC article.
References
-
- Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. - PubMed
-
- Zheng Y, Wu CX, Wu F. [Status and trends of breast cancer mortality in Chinese females.] Zhonghua Yu Fang Yi Xue Za Zhi. 2011;45:150–154. Chinese. - PubMed
-
- Blyth K, Cameron ER, Neil JC. The RUNX genes: gain or loss of function in cancer. Nat Rev Cancer. 2005;5:376–387. - PubMed
-
- Kim EJ, Kim YJ, Jeong P, Ha YS, Bae SC, Kim WJ. Methylation of the RUNX3 promoter as a potential prognostic marker for bladder tumor. J Urol. 2008;180:1141–1145. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

