Epigenetic Activation and Silencing of the Gene that Encodes IFN-γ
- PMID: 23720660
- PMCID: PMC3655339
- DOI: 10.3389/fimmu.2013.00112
Epigenetic Activation and Silencing of the Gene that Encodes IFN-γ
Abstract
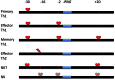
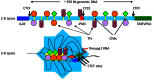
Transcriptional activation and repression of genes that are developmentally regulated or exhibit cell-type specific expression patterns is largely achieved by modifying the chromatin template at a gene locus. Complex formation of stable epigenetic histone marks, loss or gain of DNA methylation, alterations in chromosome conformation, and specific utilization of both proximal and distal transcriptional enhancers and repressors all contribute to this process. In addition, long non-coding RNAs are a new species of regulatory RNAs that either positively or negatively regulate transcription of target gene loci. IFN-γ is a pro-inflammatory cytokine with critical functions in both innate and adaptive arms of the immune system. This review focuses on our current understanding of how the chromatin template is modified at the IFNG locus during developmental processes leading to its transcriptional activation and silencing.
Keywords: CpG methylation; T helper cells; epigenetics; interferon-gamma; long non-coding RNA; natural killer T cells; natural killer cells.
Figures

Similar articles
-
Epigenetics and T helper 1 differentiation.Immunology. 2009 Mar;126(3):299-305. doi: 10.1111/j.1365-2567.2008.03026.x. Epub 2008 Dec 18. Immunology. 2009. PMID: 19178593 Free PMC article. Review.
-
Regulation of interferon-gamma during innate and adaptive immune responses.Adv Immunol. 2007;96:41-101. doi: 10.1016/S0065-2776(07)96002-2. Adv Immunol. 2007. PMID: 17981204 Review.
-
Epigenetic Control of IFN-γ Host Responses During Infection With Toxoplasma gondii.Front Immunol. 2020 Sep 25;11:581241. doi: 10.3389/fimmu.2020.581241. eCollection 2020. Front Immunol. 2020. PMID: 33072127 Free PMC article.
-
A distal region in the interferon-gamma gene is a site of epigenetic remodeling and transcriptional regulation by interleukin-2.J Biol Chem. 2004 Sep 24;279(39):41249-57. doi: 10.1074/jbc.M401168200. Epub 2004 Jul 22. J Biol Chem. 2004. PMID: 15271977
-
Sin3B mediates collagen type I gene repression by interferon gamma in vascular smooth muscle cells.Biochem Biophys Res Commun. 2014 May 2;447(2):263-70. doi: 10.1016/j.bbrc.2014.03.140. Epub 2014 Apr 4. Biochem Biophys Res Commun. 2014. PMID: 24709079
Cited by
-
UHRF1 Deficiency Inhibits Alphaherpesvirus through Inducing RIG-I-IRF3-Mediated Interferon Production.J Virol. 2023 Mar 30;97(3):e0013423. doi: 10.1128/jvi.00134-23. Epub 2023 Mar 14. J Virol. 2023. PMID: 36916938 Free PMC article.
-
Immune Biomarkers in Blood from Sarcoma Patients: A Pilot Study.Curr Oncol. 2022 Aug 5;29(8):5585-5603. doi: 10.3390/curroncol29080441. Curr Oncol. 2022. PMID: 36005179 Free PMC article.
-
Interferon-γ and systemic autoimmunity.Discov Med. 2013 Sep;16(87):123-31. Discov Med. 2013. PMID: 23998448 Free PMC article. Review.
-
Single-nucleotide methylation specifically represses type I interferon in antiviral innate immunity.J Exp Med. 2021 Mar 1;218(3):e20201798. doi: 10.1084/jem.20201798. J Exp Med. 2021. PMID: 33616624 Free PMC article.
-
TBX21-1993T/C (rs4794067) polymorphism is associated with increased risk of chronic periodontitis and increased T-bet expression in periodontal lesions, but does not significantly impact the IFN-g transcriptional level or the pattern of periodontophatic bacterial infection.Virulence. 2015;6(3):293-304. doi: 10.1080/21505594.2015.1029828. Virulence. 2015. PMID: 25832120 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

