Human papillomavirus (HPV) upregulates the cellular deubiquitinase UCHL1 to suppress the keratinocyte's innate immune response
- PMID: 23717208
- PMCID: PMC3662672
- DOI: 10.1371/journal.ppat.1003384
Human papillomavirus (HPV) upregulates the cellular deubiquitinase UCHL1 to suppress the keratinocyte's innate immune response
Abstract
Persistent infection of basal keratinocytes with high-risk human papillomavirus (hrHPV) may cause cancer. Keratinocytes are equipped with different pattern recognition receptors (PRRs) but hrHPV has developed ways to dampen their signals resulting in minimal inflammation and evasion of host immunity for sustained periods of time. To understand the mechanisms underlying hrHPV's capacity to evade immunity, we studied PRR signaling in non, newly, and persistently hrHPV-infected keratinocytes. We found that active infection with hrHPV hampered the relay of signals downstream of the PRRs to the nucleus, thereby affecting the production of type-I interferon and pro-inflammatory cytokines and chemokines. This suppression was shown to depend on hrHPV-induced expression of the cellular protein ubiquitin carboxyl-terminal hydrolase L1 (UCHL1) in keratinocytes. UCHL1 accomplished this by inhibiting tumor necrosis factor receptor-associated factor 3 (TRAF3) K63 poly-ubiquitination which lead to lower levels of TRAF3 bound to TANK-binding kinase 1 and a reduced phosphorylation of interferon regulatory factor 3. Furthermore, UCHL1 mediated the degradation of the NF-kappa-B essential modulator with as result the suppression of p65 phosphorylation and canonical NF-κB signaling. We conclude that hrHPV exploits the cellular protein UCHL1 to evade host innate immunity by suppressing PRR-induced keratinocyte-mediated production of interferons, cytokines and chemokines, which normally results in the attraction and activation of an adaptive immune response. This identifies UCHL1 as a negative regulator of PRR-induced immune responses and consequently its virus-increased expression as a strategy for hrHPV to persist.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
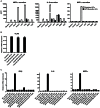
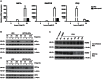

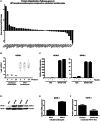
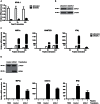


Similar articles
-
The Human Papillomavirus E6 Oncoprotein Targets USP15 and TRIM25 To Suppress RIG-I-Mediated Innate Immune Signaling.J Virol. 2018 Feb 26;92(6):e01737-17. doi: 10.1128/JVI.01737-17. Print 2018 Mar 15. J Virol. 2018. PMID: 29263274 Free PMC article.
-
Suppression of Stromal Interferon Signaling by Human Papillomavirus 16.J Virol. 2019 Sep 12;93(19):e00458-19. doi: 10.1128/JVI.00458-19. Print 2019 Oct 1. J Virol. 2019. PMID: 31292244 Free PMC article.
-
Ubiquitin-specific protease 25 regulates TLR4-dependent innate immune responses through deubiquitination of the adaptor protein TRAF3.Sci Signal. 2013 May 14;6(275):ra35. doi: 10.1126/scisignal.2003708. Sci Signal. 2013. PMID: 23674823 Free PMC article.
-
High-risk human papillomavirus targets crossroads in immune signaling.Viruses. 2015 May 21;7(5):2485-506. doi: 10.3390/v7052485. Viruses. 2015. PMID: 26008697 Free PMC article. Review.
-
Evasion of host immune defenses by human papillomavirus.Virus Res. 2017 Mar 2;231:21-33. doi: 10.1016/j.virusres.2016.11.023. Epub 2016 Nov 24. Virus Res. 2017. PMID: 27890631 Free PMC article. Review.
Cited by
-
Epidemiology, Molecular Pathogenesis, Immuno-Pathogenesis, Immune Escape Mechanisms and Vaccine Evaluation for HPV-Associated Carcinogenesis.Pathogens. 2023 Nov 23;12(12):1380. doi: 10.3390/pathogens12121380. Pathogens. 2023. PMID: 38133265 Free PMC article. Review.
-
Activation of NF-κB by human papillomavirus 16 E1 limits E1-dependent viral replication through degradation of E1.J Virol. 2015 May;89(9):5040-59. doi: 10.1128/JVI.00389-15. Epub 2015 Feb 25. J Virol. 2015. PMID: 25717108 Free PMC article.
-
Deubiquitinating Enzyme Inhibitors Block Chikungunya Virus Replication.Viruses. 2023 Feb 9;15(2):481. doi: 10.3390/v15020481. Viruses. 2023. PMID: 36851696 Free PMC article.
-
Cross-talk of cutaneous beta human papillomaviruses and the immune system: determinants of disease penetrance.Philos Trans R Soc Lond B Biol Sci. 2019 May 27;374(1773):20180287. doi: 10.1098/rstb.2018.0287. Philos Trans R Soc Lond B Biol Sci. 2019. PMID: 30955489 Free PMC article. Review.
-
Mechanisms underlying the inhibition of interferon signaling by viruses.Virulence. 2014 Feb 15;5(2):270-7. doi: 10.4161/viru.27902. Epub 2014 Feb 6. Virulence. 2014. PMID: 24504013 Free PMC article. Review.
References
-
- zur Hausen H (2002) Papillomaviruses and cancer: from basic studies to clinical application. Nat Rev Cancer 2: 342–350. - PubMed
-
- Doorbar J (2006) Molecular biology of human papillomavirus infection and cervical cancer. Clin Sci (Lond) 110: 525–541. - PubMed
-
- Frazer IH (2009) Interaction of human papillomaviruses with the host immune system: a well evolved relationship. Virology 384: 410–414. - PubMed
-
- Richardson H, Kelsall G, Tellier P, Voyer H, Abrahamowicz M, et al. (2003) The natural history of type-specific human papillomavirus infections in female university students. Cancer Epidemiol Biomarkers Prev 12: 485–490. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

