A structural chemogenomics analysis of aminergic GPCRs: lessons for histamine receptor ligand design
- PMID: 23713847
- PMCID: PMC3764853
- DOI: 10.1111/bph.12248
A structural chemogenomics analysis of aminergic GPCRs: lessons for histamine receptor ligand design
Abstract
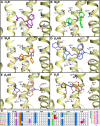
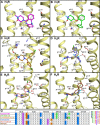
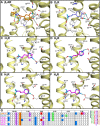
Background and purpose: Chemogenomics focuses on the discovery of new connections between chemical and biological space leading to the discovery of new protein targets and biologically active molecules. G-protein coupled receptors (GPCRs) are a particularly interesting protein family for chemogenomics studies because there is an overwhelming amount of ligand binding affinity data available. The increasing number of aminergic GPCR crystal structures now for the first time allows the integration of chemogenomics studies with high-resolution structural analyses of GPCR-ligand complexes.
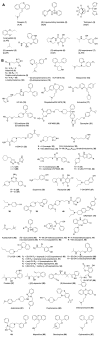
Experimental approach: In this study, we have combined ligand affinity data, receptor mutagenesis studies, and amino acid sequence analyses to high-resolution structural analyses of (hist)aminergic GPCR-ligand interactions. This integrated structural chemogenomics analysis is used to more accurately describe the molecular and structural determinants of ligand affinity and selectivity in different key binding regions of the crystallized aminergic GPCRs, and histamine receptors in particular.
Key results: Our investigations highlight interesting correlations and differences between ligand similarity and ligand binding site similarity of different aminergic receptors. Apparent discrepancies can be explained by combining detailed analysis of crystallized or predicted protein-ligand binding modes, receptor mutation studies, and ligand structure-selectivity relationships that identify local differences in essential pharmacophore features in the ligand binding sites of different receptors.
Conclusions and implications: We have performed structural chemogenomics studies that identify links between (hist)aminergic receptor ligands and their binding sites and binding modes. This knowledge can be used to identify structure-selectivity relationships that increase our understanding of ligand binding to (hist)aminergic receptors and hence can be used in future GPCR ligand discovery and design.
Keywords: GPCR; aminergic; chemical similarity; chemogenomics; crystal structures; histamine receptors; protein-ligand interactions; site-directed mutagenesis; structure-affinity relationship; transmembrane proteins.
© 2013 The British Pharmacological Society.
Figures


Similar articles
-
Aminergic GPCR-Ligand Interactions: A Chemical and Structural Map of Receptor Mutation Data.J Med Chem. 2019 Apr 25;62(8):3784-3839. doi: 10.1021/acs.jmedchem.8b00836. Epub 2018 Nov 27. J Med Chem. 2019. PMID: 30351004 Review.
-
A Structural Framework for GPCR Chemogenomics: What's In a Residue Number?Methods Mol Biol. 2018;1705:73-113. doi: 10.1007/978-1-4939-7465-8_4. Methods Mol Biol. 2018. PMID: 29188559
-
What can crystal structures of aminergic receptors tell us about designing subtype-selective ligands?Pharmacol Rev. 2015;67(1):198-213. doi: 10.1124/pr.114.009944. Pharmacol Rev. 2015. PMID: 25527701 Free PMC article. Review.
-
Combining quantum mechanical ligand conformation analysis and protein modeling to elucidate GPCR-ligand binding modes.ChemMedChem. 2013 Jan;8(1):49-53. doi: 10.1002/cmdc.201200412. Epub 2012 Nov 19. ChemMedChem. 2013. PMID: 23161844
-
Molecular determinants of ligand binding modes in the histamine H(4) receptor: linking ligand-based three-dimensional quantitative structure-activity relationship (3D-QSAR) models to in silico guided receptor mutagenesis studies.J Med Chem. 2011 Dec 8;54(23):8136-47. doi: 10.1021/jm201042n. Epub 2011 Nov 7. J Med Chem. 2011. PMID: 22003888
Cited by
-
Crystal structure of the α1B-adrenergic receptor reveals molecular determinants of selective ligand recognition.Nat Commun. 2022 Jan 19;13(1):382. doi: 10.1038/s41467-021-27911-3. Nat Commun. 2022. PMID: 35046410 Free PMC article.
-
Benchmarking of protein descriptor sets in proteochemometric modeling (part 2): modeling performance of 13 amino acid descriptor sets.J Cheminform. 2013 Sep 24;5(1):42. doi: 10.1186/1758-2946-5-42. J Cheminform. 2013. PMID: 24059743 Free PMC article.
-
Probe dependency in the determination of ligand binding kinetics at a prototypical G protein-coupled receptor.Sci Rep. 2019 May 27;9(1):7906. doi: 10.1038/s41598-019-44025-5. Sci Rep. 2019. PMID: 31133718 Free PMC article.
-
Generic GPCR residue numbers - aligning topology maps while minding the gaps.Trends Pharmacol Sci. 2015 Jan;36(1):22-31. doi: 10.1016/j.tips.2014.11.001. Epub 2014 Dec 22. Trends Pharmacol Sci. 2015. PMID: 25541108 Free PMC article. Review.
-
Comparative molecular field analysis and molecular dynamics studies of the dopamine D2 receptor antagonists without a protonatable nitrogen atom.Med Chem Res. 2018;27(4):1149-1166. doi: 10.1007/s00044-018-2137-5. Epub 2018 Feb 13. Med Chem Res. 2018. PMID: 29576721 Free PMC article.
References
-
- Ambrosio C, Molinari P, Cotecchia S, Costa T. Catechol-binding serines of beta(2)-adrenergic receptors control the equilibrium between active and inactive receptor states. Mol Pharmacol. 2000;57:198–210. - PubMed
-
- Attwood TK, Findlay JB. Fingerprinting G-protein-coupled receptors. Protein Eng. 1994;7:195–203. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

