Molecular regulation of stem cell quiescence
- PMID: 23698583
- PMCID: PMC3808888
- DOI: 10.1038/nrm3591
Molecular regulation of stem cell quiescence
Abstract
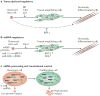
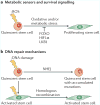
Subsets of mammalian adult stem cells reside in the quiescent state for prolonged periods of time. This state, which is reversible, has long been viewed as dormant and with minimal basal activity. Recent advances in adult stem cell isolation have provided insights into the epigenetic, transcriptional and post-transcriptional control of quiescence and suggest that quiescence is an actively maintained state in which signalling pathways are involved in maintaining a poised state that allows rapid activation. Deciphering the molecular mechanisms regulating adult stem cell quiescence will increase our understanding of tissue regeneration mechanisms and how they are dysregulated in pathological conditions and in ageing.
Figures

Similar articles
-
Mechanisms, Hallmarks, and Implications of Stem Cell Quiescence.Stem Cell Reports. 2019 Jun 11;12(6):1190-1200. doi: 10.1016/j.stemcr.2019.05.012. Stem Cell Reports. 2019. PMID: 31189093 Free PMC article. Review.
-
Molecular Regulation of Cellular Quiescence: A Perspective from Adult Stem Cells and Its Niches.Methods Mol Biol. 2018;1686:1-25. doi: 10.1007/978-1-4939-7371-2_1. Methods Mol Biol. 2018. PMID: 29030809 Review.
-
Stem Cell Quiescence: Dynamism, Restraint, and Cellular Idling.Cell Stem Cell. 2019 Feb 7;24(2):213-225. doi: 10.1016/j.stem.2019.01.001. Cell Stem Cell. 2019. PMID: 30735649 Free PMC article. Review.
-
Cellular mechanisms of somatic stem cell aging.Curr Top Dev Biol. 2014;107:405-38. doi: 10.1016/B978-0-12-416022-4.00014-7. Curr Top Dev Biol. 2014. PMID: 24439814 Free PMC article. Review.
-
Fine-tuned Rest: Unveiling the Regulatory Landscape of Adult Quiescent Neural Stem Cells.Neuroscience. 2023 Aug 10;525:26-37. doi: 10.1016/j.neuroscience.2023.07.003. Epub 2023 Jul 10. Neuroscience. 2023. PMID: 37437796 Review.
Cited by
-
The transcription factor Prox1 is essential for satellite cell differentiation and muscle fibre-type regulation.Nat Commun. 2016 Oct 12;7:13124. doi: 10.1038/ncomms13124. Nat Commun. 2016. PMID: 27731315 Free PMC article.
-
Autophagy is a key factor in maintaining the regenerative capacity of muscle stem cells by promoting quiescence and preventing senescence.Autophagy. 2016;12(4):617-8. doi: 10.1080/15548627.2016.1158373. Autophagy. 2016. PMID: 27050452 Free PMC article.
-
Mitochondria as Signaling Organelles Control Mammalian Stem Cell Fate.Cell Stem Cell. 2021 Mar 4;28(3):394-408. doi: 10.1016/j.stem.2021.02.011. Cell Stem Cell. 2021. PMID: 33667360 Free PMC article. Review.
-
p53 Inhibits Bmi-1-driven Self-Renewal and Defines Salivary Gland Cancer Stemness.Clin Cancer Res. 2022 Nov 1;28(21):4757-4770. doi: 10.1158/1078-0432.CCR-22-1357. Clin Cancer Res. 2022. PMID: 36048559 Free PMC article.
-
Odyssey of human dental pulp stem cells and their remarkable ability to survive in extremely adverse conditions.Front Physiol. 2015 Mar 26;6:99. doi: 10.3389/fphys.2015.00099. eCollection 2015. Front Physiol. 2015. PMID: 25859225 Free PMC article. No abstract available.
References
-
- Weissman IL. Stem cells: units of development, units of regeneration, and units in evolution. Cell. 2000;100:157–168. - PubMed
-
- Orford KW, Scadden DT. Deconstructing stem cell self-renewal: genetic insights into cell-cycle regulation. Nature Rev Genet. 2008;9:115–128. - PubMed
-
- Howard A, Pelc SR. Synthesis of deoxyribonucleic acid in normal and irradiated cells and its relation to chromosome breakage. Hered (Lond) [Suppl] 1953;6:261–273.
-
- Baserga R. Biochemistry of the cell cycle: a review. Cell Prolifer. 1968;1:167–191.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

