The chromatin landscape of Kaposi's sarcoma-associated herpesvirus
- PMID: 23698402
- PMCID: PMC3712311
- DOI: 10.3390/v5051346
The chromatin landscape of Kaposi's sarcoma-associated herpesvirus
Abstract
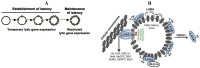
Kaposi's sarcoma-associated herpesvirus is an oncogenic γ-herpesvirus that causes latent infection in humans. In cells, the viral genome adopts a highly organized chromatin structure, which is controlled by a wide variety of cellular and viral chromatin regulatory factors. In the past few years, interrogation of the chromatinized KSHV genome by whole genome-analyzing tools revealed that the complex chromatin landscape spanning the viral genome in infected cells has important regulatory roles during the viral life cycle. This review summarizes the most recent findings regarding the role of histone modifications, histone modifying enzymes, DNA methylation, microRNAs, non-coding RNAs and the nuclear organization of the KSHV epigenome in the regulation of latent and lytic viral gene expression programs as well as their connection to KSHV-associated pathogenesis.
Figures



Similar articles
-
An atlas of chromatin landscape in KSHV-infected cells during de novo infection and reactivation.Virology. 2024 Sep;597:110146. doi: 10.1016/j.virol.2024.110146. Epub 2024 Jun 19. Virology. 2024. PMID: 38909515 Review.
-
Complex Interactions between Cohesin and CTCF in Regulation of Kaposi's Sarcoma-Associated Herpesvirus Lytic Transcription.J Virol. 2020 Jan 6;94(2):e01279-19. doi: 10.1128/JVI.01279-19. Print 2020 Jan 6. J Virol. 2020. PMID: 31666380 Free PMC article.
-
Kaposi's sarcoma-associated herpesvirus (KSHV) latency-associated nuclear antigen regulates the KSHV epigenome by association with the histone demethylase KDM3A.J Virol. 2013 Jun;87(12):6782-93. doi: 10.1128/JVI.00011-13. Epub 2013 Apr 10. J Virol. 2013. PMID: 23576503 Free PMC article.
-
Molecular biology of KSHV lytic reactivation.Viruses. 2015 Jan 14;7(1):116-53. doi: 10.3390/v7010116. Viruses. 2015. PMID: 25594835 Free PMC article. Review.
-
Epigenetic regulation of Kaposi's sarcoma-associated herpesvirus replication.Semin Cancer Biol. 2009 Jun;19(3):153-7. doi: 10.1016/j.semcancer.2009.02.010. Epub 2009 Feb 21. Semin Cancer Biol. 2009. PMID: 19429478 Free PMC article. Review.
Cited by
-
MUSASHI-Mediated Expression of JMJD3, a H3K27me3 Demethylase, Is Involved in Foamy Macrophage Generation during Mycobacterial Infection.PLoS Pathog. 2016 Aug 17;12(8):e1005814. doi: 10.1371/journal.ppat.1005814. eCollection 2016 Aug. PLoS Pathog. 2016. PMID: 27532872 Free PMC article.
-
Control of Viral Latency by Episome Maintenance Proteins.Trends Microbiol. 2020 Feb;28(2):150-162. doi: 10.1016/j.tim.2019.09.002. Epub 2019 Oct 14. Trends Microbiol. 2020. PMID: 31624007 Free PMC article. Review.
-
Biphasic euchromatin-to-heterochromatin transition on the KSHV genome following de novo infection.PLoS Pathog. 2013;9(12):e1003813. doi: 10.1371/journal.ppat.1003813. Epub 2013 Dec 19. PLoS Pathog. 2013. PMID: 24367262 Free PMC article.
-
Kaposi's Sarcoma-Associated Herpesvirus ORF66 Is Essential for Late Gene Expression and Virus Production via Interaction with ORF34.J Virol. 2020 Jan 6;94(2):e01300-19. doi: 10.1128/JVI.01300-19. Print 2020 Jan 6. J Virol. 2020. PMID: 31694948 Free PMC article.
-
Epigenetic factor siRNA screen during primary KSHV infection identifies novel host restriction factors for the lytic cycle of KSHV.PLoS Pathog. 2020 Jan 10;16(1):e1008268. doi: 10.1371/journal.ppat.1008268. eCollection 2020 Jan. PLoS Pathog. 2020. PMID: 31923286 Free PMC article.
References
-
- Cesarman E., Moore P.S., Rao P.H., Inghirami G., Knowles D.M., Chang Y. In vitro establishment and characterization of two acquired immunodeficiency syndrome-related lymphoma cell lines (BC-1 and BC-2) containing Kaposi’s sarcoma-associated herpesvirus-like (KSHV) DNA sequences. Blood. 1995;86:2708–2714. - PubMed
-
- Chang Y., Cesarman E., Pessin M.S., Lee F., Culpepper J., Knowles D.M., Moore P.S. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi’s sarcoma. Science. 1994;266:1865–1869. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P01 DE019085/DE/NIDCR NIH HHS/United States
- AI073099/AI/NIAID NIH HHS/United States
- R01 DE023926/DE/NIDCR NIH HHS/United States
- R01 CA115284/CA/NCI NIH HHS/United States
- R01 AI073099/AI/NIAID NIH HHS/United States
- R01 CA031363/CA/NCI NIH HHS/United States
- UL1 TR000130/TR/NCATS NIH HHS/United States
- R21 AI105909/AI/NIAID NIH HHS/United States
- R01 CA082057/CA/NCI NIH HHS/United States
- DE019085/DE/NIDCR NIH HHS/United States
- CA31363/CA/NCI NIH HHS/United States
- CA082057/CA/NCI NIH HHS/United States
- CA115284/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

