Unique N-linked glycosylation of CasBrE Env influences its stability, processing, and viral infectivity but not its neurotoxicity
- PMID: 23698308
- PMCID: PMC3719805
- DOI: 10.1128/JVI.00392-13
Unique N-linked glycosylation of CasBrE Env influences its stability, processing, and viral infectivity but not its neurotoxicity
Abstract
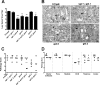
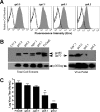
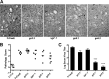
The envelope protein (Env) from the CasBrE murine leukemia virus (MLV) can cause acute spongiform neurodegeneration analogous to that induced by prions. Upon central nervous system (CNS) infection, Env is expressed as multiple isoforms owing to differential asparagine (N)-linked glycosylation. Because N-glycosylation can affect protein folding, stability, and quality control, we explored whether unique CasBrE Env glycosylation features could influence neurovirulence. CasBrE Env possesses 6/8 consensus MLV glycosylation sites (gs) but is missing gs3 and gs5 and contains a putative site (gs*). Twenty-nine mutants were generated by modifying these three sites, individually or in combination, to mimic the amino acid sequence in the nonneurovirulent Friend 57 MLV. Three basic viral phenotypes were observed: replication defective (dead; titer < 1 focus-forming unit [FFU]/ml), replication compromised (RC) (titer = 10(2) to 10(5) FFU/ml); and wild-type-like (WTL) (titer > 10(5) FFU/ml). Env protein was undetectable in dead mutants, while RC and WTL mutants showed variations in Env expression, processing, virus incorporation, virus entry, and virus spread. The newly introduced gs3 and gs5 sites were glycosylated, whereas gs* was not. Six WTL mutants tested in mice showed no clear attenuation in disease onset or severity versus controls. Furthermore, three RC viruses tested by neural stem cell (NSC)-mediated brainstem dissemination also induced acute spongiosis. Thus, while unique N-glycosylation affected structural features of Env involved in protein stability, proteolytic processing, and virus assembly and entry, these changes had minimal impact on CasBrE Env neurotoxicity. These findings suggest that the Env protein domains responsible for spongiogenesis represent highly stable elements upon which the more variable viral functional domains have evolved.
Figures




Similar articles
-
Misfolding of CasBrE SU is reversed by interactions with 4070A Env: implications for gammaretroviral neuropathogenesis.Retrovirology. 2010 Nov 5;7:93. doi: 10.1186/1742-4690-7-93. Retrovirology. 2010. PMID: 21054857 Free PMC article.
-
Retrovirus-induced spongiform neurodegeneration is mediated by unique central nervous system viral targeting and expression of env alone.J Virol. 2011 Mar;85(5):2060-78. doi: 10.1128/JVI.02210-10. Epub 2010 Dec 29. J Virol. 2011. PMID: 21191010 Free PMC article.
-
Mutational analysis of N-linked glycosylation sites of Friend murine leukemia virus envelope protein.J Virol. 1991 Oct;65(10):5323-32. doi: 10.1128/JVI.65.10.5323-5332.1991. J Virol. 1991. PMID: 1895386 Free PMC article.
-
Differential glycosylation of the Cas-Br-E env protein is associated with retrovirus-induced spongiform neurodegeneration.J Virol. 2000 Feb;74(3):1558-65. doi: 10.1128/jvi.74.3.1558-1565.2000. J Virol. 2000. PMID: 10627570 Free PMC article. Review.
-
The other transmissible spongiform encephalopathies.Rev Neurosci. 2005;16(2):159-79. doi: 10.1515/revneuro.2005.16.2.159. Rev Neurosci. 2005. PMID: 15957579 Review.
Cited by
-
Postinhibitory rebound neurons and networks are disrupted in retrovirus-induced spongiform neurodegeneration.J Neurophysiol. 2014 Aug 1;112(3):683-704. doi: 10.1152/jn.00227.2014. Epub 2014 May 14. J Neurophysiol. 2014. PMID: 25252336 Free PMC article.
References
-
- Gardner MB. 1991. Retroviral leukemia and lower motor neuron disease in wild mice: natural history, pathogenesis, and genetic resistance. Adv. Neurol. 56:473–479 - PubMed
-
- Lynch WP, Czub S, McAtee FJ, Hayes SF, Portis JL. 1991. Murine retrovirus-induced spongiform encephalopathy: productive infection of microglia and cerebellar neurons in accelerated CNS disease. Neuron 7:365–379 - PubMed
-
- Nagra RM, Masliah E, Wiley CA. 1993. Synaptic and dendritic pathology in murine retroviral encephalitis. Exp. Neurol. 124:283–288 - PubMed
-
- Czub S, Lynch WP, Czub M, Portis JL. 1994. Kinetic analysis of spongiform neurodegenerative disease induced by a highly virulent murine retrovirus. Lab. Invest. 70:711–723 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

