Abortive lytic reactivation of KSHV in CBF1/CSL deficient human B cell lines
- PMID: 23696732
- PMCID: PMC3656114
- DOI: 10.1371/journal.ppat.1003336
Abortive lytic reactivation of KSHV in CBF1/CSL deficient human B cell lines
Abstract
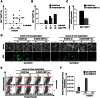
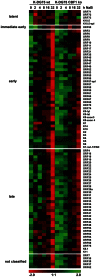
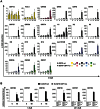
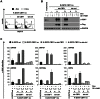
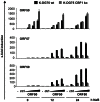
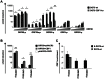
Since Kaposi's sarcoma associated herpesvirus (KSHV) establishes a persistent infection in human B cells, B cells are a critical compartment for viral pathogenesis. RTA, the replication and transcription activator of KSHV, can either directly bind to DNA or use cellular DNA binding factors including CBF1/CSL as DNA adaptors. In addition, the viral factors LANA1 and vIRF4 are known to bind to CBF1/CSL and modulate RTA activity. To analyze the contribution of CBF1/CSL to reactivation in human B cells, we have successfully infected DG75 and DG75 CBF1/CSL knock-out cell lines with recombinant KSHV.219 and selected for viral maintenance by selective medium. Both lines maintained the virus irrespective of their CBF1/CSL status. Viral reactivation could be initiated in both B cell lines but viral genome replication was attenuated in CBF1/CSL deficient lines, which also failed to produce detectable levels of infectious virus. Induction of immediate early, early and late viral genes was impaired in CBF1/CSL deficient cells at multiple stages of the reactivation process but could be restored to wild-type levels by reintroduction of CBF1/CSL. To identify additional viral RTA target genes, which are directly controlled by CBF1/CSL, we analyzed promoters of a selected subset of viral genes. We show that the induction of the late viral genes ORF29a and ORF65 by RTA is strongly enhanced by CBF1/CSL. Orthologs of ORF29a in other herpesviruses are part of the terminase complex required for viral packaging. ORF65 encodes the small capsid protein essential for capsid shell assembly. Our study demonstrates for the first time that in human B cells viral replication can be initiated in the absence of CBF1/CSL but the reactivation process is severely attenuated at all stages and does not lead to virion production. Thus, CBF1/CSL acts as a global hub which is used by the virus to coordinate the lytic cascade.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

Similar articles
-
Genome-Wide Identification of Direct RTA Targets Reveals Key Host Factors for Kaposi's Sarcoma-Associated Herpesvirus Lytic Reactivation.J Virol. 2019 Feb 19;93(5):e01978-18. doi: 10.1128/JVI.01978-18. Print 2019 Mar 1. J Virol. 2019. PMID: 30541837 Free PMC article.
-
Kaposi's sarcoma-associated herpesvirus viral interferon regulatory factor 4 (vIRF4/K10) is a novel interaction partner of CSL/CBF1, the major downstream effector of Notch signaling.J Virol. 2010 Dec;84(23):12255-64. doi: 10.1128/JVI.01484-10. Epub 2010 Sep 22. J Virol. 2010. PMID: 20861242 Free PMC article.
-
Wide-scale use of Notch signaling factor CSL/RBP-Jkappa in RTA-mediated activation of Kaposi's sarcoma-associated herpesvirus lytic genes.J Virol. 2010 Feb;84(3):1334-47. doi: 10.1128/JVI.01301-09. Epub 2009 Nov 11. J Virol. 2010. PMID: 19906914 Free PMC article.
-
Reactivation and Lytic Replication of Kaposi's Sarcoma-Associated Herpesvirus: An Update.Front Microbiol. 2017 Apr 20;8:613. doi: 10.3389/fmicb.2017.00613. eCollection 2017. Front Microbiol. 2017. PMID: 28473805 Free PMC article. Review.
-
Molecular biology of KSHV lytic reactivation.Viruses. 2015 Jan 14;7(1):116-53. doi: 10.3390/v7010116. Viruses. 2015. PMID: 25594835 Free PMC article. Review.
Cited by
-
Kaposi sarcoma herpesvirus pathogenesis.Philos Trans R Soc Lond B Biol Sci. 2017 Oct 19;372(1732):20160275. doi: 10.1098/rstb.2016.0275. Philos Trans R Soc Lond B Biol Sci. 2017. PMID: 28893942 Free PMC article. Review.
-
The cellular Notch1 protein promotes KSHV reactivation in an Rta-dependent manner.J Virol. 2024 Aug 20;98(8):e0078824. doi: 10.1128/jvi.00788-24. Epub 2024 Jul 8. J Virol. 2024. PMID: 38975769 Free PMC article.
-
KSHV and the Role of Notch Receptor Dysregulation in Disease Progression.Pathogens. 2017 Aug 4;6(3):34. doi: 10.3390/pathogens6030034. Pathogens. 2017. PMID: 28777778 Free PMC article. Review.
-
Herpesviruses dUTPases: A New Family of Pathogen-Associated Molecular Pattern (PAMP) Proteins with Implications for Human Disease.Pathogens. 2016 Dec 28;6(1):2. doi: 10.3390/pathogens6010002. Pathogens. 2016. PMID: 28036046 Free PMC article. Review.
-
MicroRNA let-7 and viral infections: focus on mechanisms of action.Cell Mol Biol Lett. 2022 Feb 14;27(1):14. doi: 10.1186/s11658-022-00317-9. Cell Mol Biol Lett. 2022. PMID: 35164678 Free PMC article. Review.
References
-
- Cesarman E, Moore PS, Rao PH, Inghirami G, Knowles DM, et al. (1995) In vitro establishment and characterization of two acquired immunodeficiency syndrome-related lymphoma cell lines (BC-1 and BC-2) containing Kaposi's sarcoma-associated herpesvirus-like (KSHV) DNA sequences. Blood 86: 2708–2714. - PubMed
-
- Ambroziak JA, Blackbourn DJ, Herndier BG, Glogau RG, Gullett JH, et al. (1995) Herpes-like sequences in HIV-infected and uninfected Kaposi's sarcoma patients. Science 268: 582–583. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

