Subtelomeric hotspots of aberrant 5-hydroxymethylcytosine-mediated epigenetic modifications during reprogramming to pluripotency
- PMID: 23685628
- PMCID: PMC3998089
- DOI: 10.1038/ncb2748
Subtelomeric hotspots of aberrant 5-hydroxymethylcytosine-mediated epigenetic modifications during reprogramming to pluripotency
Abstract
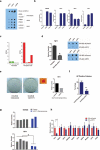
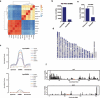
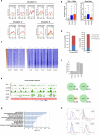
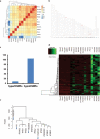
Mammalian somatic cells can be directly reprogrammed into induced pluripotent stem cells (iPSCs) by introducing defined sets of transcription factors. Somatic cell reprogramming involves epigenomic reconfiguration, conferring iPSCs with characteristics similar to embryonic stem cells (ESCs). Human ESCs (hESCs) contain 5-hydroxymethylcytosine (5hmC), which is generated through the oxidation of 5-methylcytosine by the TET enzyme family. Here we show that 5hmC levels increase significantly during reprogramming to human iPSCs mainly owing to TET1 activation, and this hydroxymethylation change is critical for optimal epigenetic reprogramming, but does not compromise primed pluripotency. Compared with hESCs, we find that iPSCs tend to form large-scale (100 kb-1.3 Mb) aberrant reprogramming hotspots in subtelomeric regions, most of which exhibit incomplete hydroxymethylation on CG sites. Strikingly, these 5hmC aberrant hotspots largely coincide (~80%) with aberrant iPSC-ESC non-CG methylation regions. Our results suggest that TET1-mediated 5hmC modification could contribute to the epigenetic variation of iPSCs and iPSC-hESC differences.
Figures

Comment in
-
-Oh no! hiPSCs misplace their 5hmCs.Cell Stem Cell. 2013 Jul 3;13(1):10-1. doi: 10.1016/j.stem.2013.06.007. Cell Stem Cell. 2013. PMID: 23827705 Free PMC article.
Similar articles
-
Replacement of Oct4 by Tet1 during iPSC induction reveals an important role of DNA methylation and hydroxymethylation in reprogramming.Cell Stem Cell. 2013 Apr 4;12(4):453-69. doi: 10.1016/j.stem.2013.02.005. Epub 2013 Mar 14. Cell Stem Cell. 2013. PMID: 23499384
-
Early-stage epigenetic modification during somatic cell reprogramming by Parp1 and Tet2.Nature. 2012 Aug 30;488(7413):652-5. doi: 10.1038/nature11333. Nature. 2012. PMID: 22902501 Free PMC article.
-
Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA.Science. 2011 Sep 2;333(6047):1303-7. doi: 10.1126/science.1210944. Epub 2011 Aug 4. Science. 2011. PMID: 21817016 Free PMC article.
-
Tet family of 5-methylcytosine dioxygenases in mammalian development.J Hum Genet. 2013 Jul;58(7):421-7. doi: 10.1038/jhg.2013.63. Epub 2013 May 30. J Hum Genet. 2013. PMID: 23719188 Free PMC article. Review.
-
DNA demethylation, Tet proteins and 5-hydroxymethylcytosine in epigenetic reprogramming: an emerging complex story.Genomics. 2014 Nov;104(5):324-33. doi: 10.1016/j.ygeno.2014.08.012. Epub 2014 Aug 27. Genomics. 2014. PMID: 25173569 Review.
Cited by
-
DNA methylation and hydroxymethylation in stem cells.Cell Biochem Funct. 2015 Jun;33(4):161-73. doi: 10.1002/cbf.3101. Epub 2015 Mar 16. Cell Biochem Funct. 2015. PMID: 25776144 Free PMC article. Review.
-
Concise review: drug discovery in the age of the induced pluripotent stem cell.Stem Cells Transl Med. 2014 Apr;3(4):500-9. doi: 10.5966/sctm.2013-0162. Epub 2014 Feb 3. Stem Cells Transl Med. 2014. PMID: 24493856 Free PMC article. Review.
-
Molecules and mechanisms controlling the active DNA demethylation of the mammalian zygotic genome.Protein Cell. 2014 Nov;5(11):827-36. doi: 10.1007/s13238-014-0095-3. Epub 2014 Aug 26. Protein Cell. 2014. PMID: 25152302 Free PMC article. Review.
-
Cancer-like epigenetic derangements of human pluripotent stem cells and their impact on applications in regeneration and repair.Curr Opin Genet Dev. 2014 Oct;28:43-9. doi: 10.1016/j.gde.2014.09.008. Epub 2014 Oct 14. Curr Opin Genet Dev. 2014. PMID: 25461449 Free PMC article. Review.
-
Methylation of subtelomeric repeat D4Z4 in peripheral blood leukocytes is associated with biochemical recurrence in localized prostate cancer patients.Carcinogenesis. 2017 Aug 1;38(8):821-826. doi: 10.1093/carcin/bgx064. Carcinogenesis. 2017. PMID: 28854562 Free PMC article.
References
-
- Takahashi K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. - PubMed
-
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. - PubMed
-
- Yu J, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
- P30 HD003352/HD/NICHD NIH HHS/United States
- R00 DE021442/DE/NIDCR NIH HHS/United States
- R01 MH092902/MH/NIMH NIH HHS/United States
- K99 DE021442/DE/NIDCR NIH HHS/United States
- P30 HD024064/HD/NICHD NIH HHS/United States
- P50 AG025688/AG/NIA NIH HHS/United States
- R01 NS051630/NS/NINDS NIH HHS/United States
- DP3 DK094346/DK/NIDDK NIH HHS/United States
- NS079625/NS/NINDS NIH HHS/United States
- HD24064/HD/NICHD NIH HHS/United States
- R01 NS079625/NS/NINDS NIH HHS/United States
- HD073162/HD/NICHD NIH HHS/United States
- R21 HD073162/HD/NICHD NIH HHS/United States
- MH089606/MH/NIMH NIH HHS/United States
- T32 MH087977/MH/NIMH NIH HHS/United States
- R01 MH102690/MH/NIMH NIH HHS/United States
- R01 HD068730/HD/NICHD NIH HHS/United States
- R01 MH089606/MH/NIMH NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

