Comparative analysis of the repertoire of G protein-coupled receptors of three species of the fungal genus Trichoderma
- PMID: 23679152
- PMCID: PMC3664084
- DOI: 10.1186/1471-2180-13-108
Comparative analysis of the repertoire of G protein-coupled receptors of three species of the fungal genus Trichoderma
Abstract
Background: Eukaryotic organisms employ cell surface receptors such as the seven-transmembrane G protein-coupled receptors (GPCRs) as sensors to connect to the environment. GPCRs react to a variety of extracellular cues and are considered to play central roles in the signal transduction in fungi. Several species of the filamentous ascomycete Trichoderma are potent mycoparasites, i.e. can attack and parasitize other fungi, which turns them into successful bio-fungicides for the protection of plants against fungal phytopathogens. The identification and characterization of GPCRs will provide insights into how Trichoderma communicates with its environment and senses the presence of host fungi.
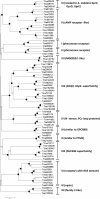
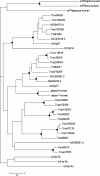
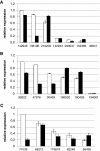
Results: We mined the recently published genomes of the two mycoparasitic biocontrol agents Trichoderma atroviride and Trichoderma virens and compared the identified GPCR-like proteins to those of the saprophyte Trichoderma reesei. Phylogenetic analyses resulted in 14 classes and revealed differences not only among the three Trichoderma species but also between Trichoderma and other fungi. The class comprising proteins of the PAQR family was significantly expanded both in Trichoderma compared to other fungi as well as in the two mycoparasites compared to T. reesei. Expression analysis of the PAQR-encoding genes of the three Trichoderma species revealed that all except one were actually transcribed. Furthermore, the class of receptors with a DUF300 domain was expanded in T. atroviride, and T. virens showed an expansion of PTH11-like receptors compared to T. atroviride and T. reesei.
Conclusions: Comparative genome analyses of three Trichoderma species revealed a great diversity of putative GPCRs with genus- and species- specific differences. The expansion of certain classes in the mycoparasites T. atroviride and T. virens is likely to reflect the capability of these fungi to establish various ecological niches and interactions with other organisms such as fungi and plants. These GPCRs consequently represent interesting candidates for future research on the mechanisms underlying mycoparasitism and biocontrol.
Figures
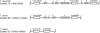
Similar articles
-
The Genomes of Three Uneven Siblings: Footprints of the Lifestyles of Three Trichoderma Species.Microbiol Mol Biol Rev. 2016 Feb 10;80(1):205-327. doi: 10.1128/MMBR.00040-15. Print 2016 Mar. Microbiol Mol Biol Rev. 2016. PMID: 26864432 Free PMC article. Review.
-
Trichoderma G protein-coupled receptors: functional characterisation of a cAMP receptor-like protein from Trichoderma atroviride.Curr Genet. 2008 Dec;54(6):283-99. doi: 10.1007/s00294-008-0217-7. Epub 2008 Oct 3. Curr Genet. 2008. PMID: 18836726 Free PMC article.
-
Phylogenomic analysis of polyketide synthase-encoding genes in Trichoderma.Microbiology (Reading). 2012 Jan;158(Pt 1):147-154. doi: 10.1099/mic.0.053462-0. Epub 2011 Nov 17. Microbiology (Reading). 2012. PMID: 22096146
-
A comparative cell wall analysis of Trichoderma spp. confirms a conserved polysaccharide scaffold and suggests an important role for chitosan in mycoparasitism.Microbiol Spectr. 2024 Aug 6;12(8):e0349523. doi: 10.1128/spectrum.03495-23. Epub 2024 Jun 25. Microbiol Spectr. 2024. PMID: 38916333 Free PMC article.
-
Novel traits of Trichoderma predicted through the analysis of its secretome.FEMS Microbiol Lett. 2012 Dec;337(1):1-9. doi: 10.1111/j.1574-6968.2012.02665.x. Epub 2012 Sep 18. FEMS Microbiol Lett. 2012. PMID: 22924408 Free PMC article. Review.
Cited by
-
Identification of a key G-protein coupled receptor in mediating appressorium formation and fungal virulence against insects.Sci China Life Sci. 2021 Mar;64(3):466-477. doi: 10.1007/s11427-020-1763-1. Epub 2020 Jul 23. Sci China Life Sci. 2021. PMID: 32712834
-
Global survey of canonical Aspergillus flavus G protein-coupled receptors.mBio. 2014 Oct 14;5(5):e01501-14. doi: 10.1128/mBio.01501-14. mBio. 2014. PMID: 25316696 Free PMC article.
-
GPCRs from fusarium graminearum detection, modeling and virtual screening - the search for new routes to control head blight disease.BMC Bioinformatics. 2016 Dec 15;17(Suppl 18):463. doi: 10.1186/s12859-016-1342-9. BMC Bioinformatics. 2016. PMID: 28105916 Free PMC article.
-
cAMP activates calcium signalling via phospholipase C to regulate cellulase production in the filamentous fungus Trichoderma reesei.Biotechnol Biofuels. 2021 Mar 8;14(1):62. doi: 10.1186/s13068-021-01914-0. Biotechnol Biofuels. 2021. PMID: 33685506 Free PMC article.
-
The Genomes of Three Uneven Siblings: Footprints of the Lifestyles of Three Trichoderma Species.Microbiol Mol Biol Rev. 2016 Feb 10;80(1):205-327. doi: 10.1128/MMBR.00040-15. Print 2016 Mar. Microbiol Mol Biol Rev. 2016. PMID: 26864432 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

