Enhancement of proteolytic activity of a thermostable papain-like protease by structure-based rational design
- PMID: 23671614
- PMCID: PMC3643963
- DOI: 10.1371/journal.pone.0062619
Enhancement of proteolytic activity of a thermostable papain-like protease by structure-based rational design
Abstract
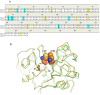
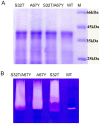
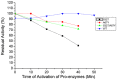
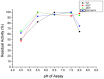
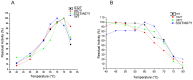
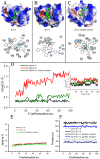
Ervatamins (A, B and C) are papain-like cysteine proteases from the plant Ervatamia coronaria. Among Ervatamins, Ervatamin-C is a thermostable protease, but it shows lower catalytic efficiency. In contrast, Ervatamin-A which has a high amino acid sequence identity (∼90%) and structural homology (Cα rmsd 0.4 Å) with Ervatamin-C, has much higher catalytic efficiency (∼57 times). From the structural comparison of Ervatamin-A and -C, two residues Thr32 and Tyr67 in the catalytic cleft of Ervatamin-A have been identified whose contributions for higher activity of Ervatamin-A are established in our earlier studies. In this study, these two residues have been introduced in Ervatamin-C by site directed mutagenesis to enhance the catalytic efficiency of the thermostable protease. Two single mutants (S32T and A67Y) and one double mutant (S32T/A67Y) of Ervatamin-C have been generated and characterized. All the three mutants show ∼ 8 times higher catalytic efficiency (k cat/K m) than the wild-type. The thermostability of all the three mutant enzymes remained unchanged. The double mutant does not achieve the catalytic efficiency of the template enzyme Ervatamin-A. By modeling the structure of the double mutant and probing the role of active site residues by docking a substrate, the mechanistic insights of higher activity of the mutant protease have been addressed. The in-silico study demonstrates that the residues beyond the catalytic cleft also influence the substrate binding and positioning of the substrate at the catalytic centre, thus controlling the catalytic efficiency of an enzyme.
Conflict of interest statement
Figures

Similar articles
-
Structural insights into the substrate specificity and activity of ervatamins, the papain-like cysteine proteases from a tropical plant, Ervatamia coronaria.FEBS J. 2008 Feb;275(3):421-34. doi: 10.1111/j.1742-4658.2007.06211.x. Epub 2007 Dec 19. FEBS J. 2008. PMID: 18167146
-
Structural basis of the unusual stability and substrate specificity of ervatamin C, a plant cysteine protease from Ervatamia coronaria.Biochemistry. 2004 Feb 17;43(6):1532-40. doi: 10.1021/bi0357659. Biochemistry. 2004. PMID: 14769029
-
C-Terminal extension of a plant cysteine protease modulates proteolytic activity through a partial inhibitory mechanism.FEBS J. 2011 Sep;278(17):3012-24. doi: 10.1111/j.1742-4658.2011.08221.x. Epub 2011 Jul 18. FEBS J. 2011. PMID: 21707922
-
Papain-like lysosomal cysteine proteases and their inhibitors: drug discovery targets?Biochem Soc Symp. 2003;(70):15-30. doi: 10.1042/bss0700015. Biochem Soc Symp. 2003. PMID: 14587279 Review.
-
Plant asparaginyl endopeptidases and their structural determinants of function.Biochem Soc Trans. 2021 Apr 30;49(2):965-976. doi: 10.1042/BST20200908. Biochem Soc Trans. 2021. PMID: 33666219 Free PMC article. Review.
Cited by
-
Structure determinants defining the specificity of papain-like cysteine proteases.Comput Struct Biotechnol J. 2022 Nov 24;20:6552-6569. doi: 10.1016/j.csbj.2022.11.040. eCollection 2022. Comput Struct Biotechnol J. 2022. PMID: 36467578 Free PMC article. Review.
References
-
- Grudkowska M, Zagdanska B (2004) Multifunctional role of plant cysteine proteinases. Acta Biochim Pol 51: 609–624. - PubMed
-
- Barrett AJ, Rawlings ND, Woessner JF (1998) Handbook of proteolytic enzymes. second ed. London: Academic Press.
-
- Baker PJ, Numata K (2012) Chemoenzymatic synthesis of 1 Poly (L-alanine) in aqueous environment. Biomacromolecules 13: 947–951. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

