Obstacles may facilitate and direct DNA search by proteins
- PMID: 23663847
- PMCID: PMC3647173
- DOI: 10.1016/j.bpj.2013.03.030
Obstacles may facilitate and direct DNA search by proteins
Abstract
DNA recognition by DNA-binding proteins (DBPs), which is a pivotal event in most gene regulatory processes, is often preceded by an extensive search for the correct site. A facilitated diffusion process in which a DBP combines three-dimensional diffusion in solution with one-dimensional sliding along DNA has been suggested to explain how proteins can locate their target sites on DNA much faster than predicted by three-dimensional diffusion alone. Although experimental and theoretical studies have recently advanced understanding of the biophysical principles underlying the search mechanism, the process under in vivo cellular conditions is poorly understood. In this study, we used various computational approaches to explore how the presence of obstacle proteins on the DNA influences search efficiency. At a low obstacle occupancy (i.e., when few obstacles occupy sites on the DNA), sliding by the searching DBP may be confined, which may impair search efficiency. The obstacles, however, can be bypassed during hopping events, and the number of bypasses is larger for higher obstacle occupancies. Dynamism on the part of the obstacles may even further facilitate search kinetics. Our study shows that the nature and efficiency of the search process may be governed not only by the intrinsic properties of the DBP and the salt concentration of the medium, but also by the in vivo association of DNA with other macromolecular obstacles, their location, and occupancy.
Copyright © 2013 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures
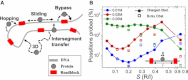
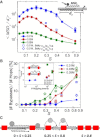

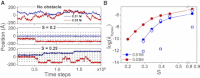
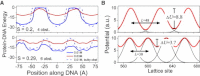
Comment in
-
Please, get in my way so that I can be more efficient!Biophys J. 2013 May 7;104(9):1842-3. doi: 10.1016/j.bpj.2013.03.042. Biophys J. 2013. PMID: 23663825 Free PMC article. No abstract available.
Similar articles
-
Searching target sites on DNA by proteins: Role of DNA dynamics under confinement.Nucleic Acids Res. 2015 Oct 30;43(19):9176-86. doi: 10.1093/nar/gkv931. Epub 2015 Sep 22. Nucleic Acids Res. 2015. PMID: 26400158 Free PMC article.
-
Theoretical and computational modeling of target-site search kinetics in vitro and in vivo.Biophys J. 2011 Aug 17;101(4):856-65. doi: 10.1016/j.bpj.2011.06.066. Biophys J. 2011. PMID: 21843476 Free PMC article.
-
Molecular crowding effect on dynamics of DNA-binding proteins search for their targets.J Chem Phys. 2014 Dec 14;141(22):225102. doi: 10.1063/1.4903505. J Chem Phys. 2014. PMID: 25494769
-
Facilitated diffusion in chromatin lattices: mechanistic diversity and regulatory potential.Mol Microbiol. 2005 Aug;57(4):889-99. doi: 10.1111/j.1365-2958.2005.04707.x. Mol Microbiol. 2005. PMID: 16091032 Review.
-
Physics of protein-DNA interactions: mechanisms of facilitated target search.Phys Chem Chem Phys. 2011 Feb 14;13(6):2088-95. doi: 10.1039/c0cp01966f. Epub 2010 Nov 29. Phys Chem Chem Phys. 2011. PMID: 21113556 Review.
Cited by
-
Search by proteins for their DNA target site: 1. The effect of DNA conformation on protein sliding.Nucleic Acids Res. 2014 Nov 10;42(20):12404-14. doi: 10.1093/nar/gku932. Epub 2014 Oct 16. Nucleic Acids Res. 2014. PMID: 25324308 Free PMC article.
-
Mechanisms of Protein Search for Targets on DNA: Theoretical Insights.Molecules. 2018 Aug 22;23(9):2106. doi: 10.3390/molecules23092106. Molecules. 2018. PMID: 30131459 Free PMC article.
-
Optimal Length of Conformational Transition Region in Protein Search for Targets on DNA.J Phys Chem Lett. 2017 Sep 7;8(17):4049-4054. doi: 10.1021/acs.jpclett.7b01750. Epub 2017 Aug 15. J Phys Chem Lett. 2017. PMID: 28796515 Free PMC article.
-
Structural Basis of Enhanced Facilitated Diffusion of DNA-Binding Protein in Crowded Cellular Milieu.Biophys J. 2020 Jan 21;118(2):505-517. doi: 10.1016/j.bpj.2019.11.3388. Epub 2019 Nov 29. Biophys J. 2020. PMID: 31862109 Free PMC article.
-
Real sequence effects on the search dynamics of transcription factors on DNA.Sci Rep. 2015 Jul 8;5:10072. doi: 10.1038/srep10072. Sci Rep. 2015. PMID: 26154484 Free PMC article.
References
-
- Zakrzewska K., Lavery R. Towards a molecular view of transcriptional control. Curr. Opin. Struct. Biol. 2012;22:160–167. - PubMed
-
- Riggs A.D., Bourgeois S., Cohn M. The lac repressor-operator interaction. 3. Kinetic studies. J. Mol. Biol. 1970;53:401–417. - PubMed
-
- Berg O.G., Winter R.B., von Hippel P.H. Diffusion-driven mechanisms of protein translocation on nucleic acids. 1. Models and theory. Biochemistry. 1981;20:6929–6948. - PubMed
-
- von Hippel P.H., Berg O.G. Facilitated target location in biological systems. J. Biol. Chem. 1989;264:675–678. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

