Control of DNA minor groove width and Fis protein binding by the purine 2-amino group
- PMID: 23661683
- PMCID: PMC3711457
- DOI: 10.1093/nar/gkt357
Control of DNA minor groove width and Fis protein binding by the purine 2-amino group
Abstract
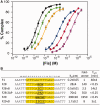
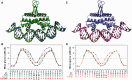
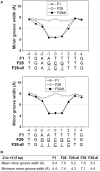
The width of the DNA minor groove varies with sequence and can be a major determinant of DNA shape recognition by proteins. For example, the minor groove within the center of the Fis-DNA complex narrows to about half the mean minor groove width of canonical B-form DNA to fit onto the protein surface. G/C base pairs within this segment, which is not contacted by the Fis protein, reduce binding affinities up to 2000-fold over A/T-rich sequences. We show here through multiple X-ray structures and binding properties of Fis-DNA complexes containing base analogs that the 2-amino group on guanine is the primary molecular determinant controlling minor groove widths. Molecular dynamics simulations of free-DNA targets with canonical and modified bases further demonstrate that sequence-dependent narrowing of minor groove widths is modulated almost entirely by the presence of purine 2-amino groups. We also provide evidence that protein-mediated phosphate neutralization facilitates minor groove compression and is particularly important for binding to non-optimally shaped DNA duplexes.
Figures


Similar articles
-
DNA Sequence Determinants Controlling Affinity, Stability and Shape of DNA Complexes Bound by the Nucleoid Protein Fis.PLoS One. 2016 Mar 9;11(3):e0150189. doi: 10.1371/journal.pone.0150189. eCollection 2016. PLoS One. 2016. PMID: 26959646 Free PMC article.
-
The shape of the DNA minor groove directs binding by the DNA-bending protein Fis.Genes Dev. 2010 Apr 15;24(8):814-26. doi: 10.1101/gad.1900610. Genes Dev. 2010. PMID: 20395367 Free PMC article.
-
Cooperative DNA binding by proteins through DNA shape complementarity.Nucleic Acids Res. 2019 Sep 19;47(16):8874-8887. doi: 10.1093/nar/gkz642. Nucleic Acids Res. 2019. PMID: 31616952 Free PMC article.
-
Binding to the DNA Minor Groove by Heterocyclic Dications: from AT Specific to GC Recognition Compounds.Curr Protoc. 2023 Apr;3(4):e729. doi: 10.1002/cpz1.729. Curr Protoc. 2023. PMID: 37071034 Review.
-
DNA recognition by beta-sheets.Biopolymers. 1997;44(4):335-59. doi: 10.1002/(SICI)1097-0282(1997)44:4<335::AID-BIP3>3.0.CO;2-R. Biopolymers. 1997. PMID: 9782775 Review.
Cited by
-
Temporal hierarchy of gene expression mediated by transcription factor binding affinity and activation dynamics.mBio. 2015 May 26;6(3):e00686-15. doi: 10.1128/mBio.00686-15. mBio. 2015. PMID: 26015501 Free PMC article.
-
Structural Basis for the Inverted Repeat Preferences of mariner Transposases.J Biol Chem. 2015 May 22;290(21):13531-40. doi: 10.1074/jbc.M115.636704. Epub 2015 Apr 13. J Biol Chem. 2015. PMID: 25869132 Free PMC article.
-
The unusual monomer recognition of guanine-containing mixed sequence DNA by a dithiophene heterocyclic diamidine.Biochemistry. 2014 Feb 25;53(7):1218-27. doi: 10.1021/bi401582t. Epub 2014 Feb 13. Biochemistry. 2014. PMID: 24495039 Free PMC article.
-
TFBSshape: a motif database for DNA shape features of transcription factor binding sites.Nucleic Acids Res. 2014 Jan;42(Database issue):D148-55. doi: 10.1093/nar/gkt1087. Epub 2013 Nov 7. Nucleic Acids Res. 2014. PMID: 24214955 Free PMC article.
-
DNA Sequence Determinants Controlling Affinity, Stability and Shape of DNA Complexes Bound by the Nucleoid Protein Fis.PLoS One. 2016 Mar 9;11(3):e0150189. doi: 10.1371/journal.pone.0150189. eCollection 2016. PLoS One. 2016. PMID: 26959646 Free PMC article.
References
-
- Otwinowski Z, Schevitz RW, Zhang RG, Lawson CL, Joachimiak A, Marmorstein RQ, Luisi BF, Sigler PB. Crystal structure of Trp repressor/operator complex at atomic resolution. Nature. 1988;335:321–329. - PubMed
-
- Johnson RC, Stella S, Heiss JK. In: Protein-Nucleic Acid Interactions. Rice PA, Correll CC, editors. RSC Press: Cambridge, UK; 2008. pp. 176–220.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

