Epigenetics of the antibody response
- PMID: 23643790
- PMCID: PMC3744588
- DOI: 10.1016/j.it.2013.03.006
Epigenetics of the antibody response
Abstract
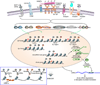
Epigenetic marks, such as DNA methylation, histone post-translational modifications and miRNAs, are induced in B cells by the same stimuli that drive the antibody response. They play major roles in regulating somatic hypermutation (SHM), class switch DNA recombination (CSR), and differentiation to plasma cells or long-lived memory B cells. Histone modifications target the CSR and, possibly, SHM machinery to the immunoglobulin locus; they together with DNA methylation and miRNAs modulate the expression of critical elements of that machinery, such as activation-induced cytidine deaminase (AID), as well as factors central to plasma cell differentiation, such as B lymphocyte-induced maturation protein-1 (Blimp-1). These inducible B cell-intrinsic epigenetic marks instruct the maturation of antibody responses. Their dysregulation plays an important role in aberrant antibody responses to foreign antigens, such as those of microbial pathogens, and self-antigens, such as those targeted in autoimmunity, and B cell neoplasia.
Keywords: AID; B cell; Blimp-1; CSR; SHM; antibody; autoimmunity; epigenetic; immunoglobulin; memory B cell; neoplasia; plasma cell.
Copyright © 2013 Elsevier Ltd. All rights reserved.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


Similar articles
-
B cell Rab7 mediates induction of activation-induced cytidine deaminase expression and class-switching in T-dependent and T-independent antibody responses.J Immunol. 2015 Apr 1;194(7):3065-78. doi: 10.4049/jimmunol.1401896. Epub 2015 Mar 4. J Immunol. 2015. PMID: 25740947 Free PMC article.
-
Epigenetics of Peripheral B-Cell Differentiation and the Antibody Response.Front Immunol. 2015 Dec 14;6:631. doi: 10.3389/fimmu.2015.00631. eCollection 2015. Front Immunol. 2015. PMID: 26697022 Free PMC article. Review.
-
Histone deacetylase inhibitors upregulate B cell microRNAs that silence AID and Blimp-1 expression for epigenetic modulation of antibody and autoantibody responses.J Immunol. 2014 Dec 15;193(12):5933-50. doi: 10.4049/jimmunol.1401702. Epub 2014 Nov 12. J Immunol. 2014. PMID: 25392531 Free PMC article.
-
Epigenetics of the antibody and autoantibody response.Curr Opin Immunol. 2020 Dec;67:75-86. doi: 10.1016/j.coi.2020.09.004. Epub 2020 Nov 8. Curr Opin Immunol. 2020. PMID: 33176228 Free PMC article. Review.
-
Independent Roles of Switching and Hypermutation in the Development and Persistence of B Lymphocyte Memory.Immunity. 2016 Apr 19;44(4):769-81. doi: 10.1016/j.immuni.2016.01.011. Epub 2016 Mar 2. Immunity. 2016. PMID: 26944202 Free PMC article.
Cited by
-
Regulation of Aicda expression and AID activity.Autoimmunity. 2013 Mar;46(2):83-101. doi: 10.3109/08916934.2012.749244. Epub 2013 Jan 17. Autoimmunity. 2013. PMID: 23181381 Free PMC article. Review.
-
Behavioral and immunohistochemical characterization of rapid reconditioning following extinction of contextual fear.Learn Mem. 2019 Sep 16;26(10):1-16. doi: 10.1101/lm.048439.118. Print 2019 Oct. Learn Mem. 2019. PMID: 31527183 Free PMC article.
-
Genome-Wide Analysis Reveals Selective Modulation of microRNAs and mRNAs by Histone Deacetylase Inhibitor in B Cells Induced to Undergo Class-Switch DNA Recombination and Plasma Cell Differentiation.Front Immunol. 2015 Dec 14;6:627. doi: 10.3389/fimmu.2015.00627. eCollection 2015. Front Immunol. 2015. PMID: 26697020 Free PMC article.
-
B cell Rab7 mediates induction of activation-induced cytidine deaminase expression and class-switching in T-dependent and T-independent antibody responses.J Immunol. 2015 Apr 1;194(7):3065-78. doi: 10.4049/jimmunol.1401896. Epub 2015 Mar 4. J Immunol. 2015. PMID: 25740947 Free PMC article.
-
Generating and repairing genetically programmed DNA breaks during immunoglobulin class switch recombination.F1000Res. 2018 Apr 13;7:458. doi: 10.12688/f1000research.13247.1. eCollection 2018. F1000Res. 2018. PMID: 29744038 Free PMC article. Review.
References
-
- Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 2012;13:484–492. - PubMed
-
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. - PubMed
-
- Esteller M. Non-coding RNAs in human disease. Nat. Rev. Genet. 2011;12:861–874. - PubMed
-
- Schatz DG, Ji Y. Recombination centres and the orchestration of V(D)J recombination. Nat. Rev. Immunol. 2011;11:251–263. - PubMed
-
- Shaknovich R, Cerchietti L, Tsikitas L, Kormaksson M, De S, Figueroa ME, Ballon G, Yang SN, Weinhold N, Reimers M, Clozel T, Luttrop K, Ekstrom TJ, Frank J, Vasanthakumar A, Godley LA, Michor F, Elemento O, Melnick A. DNA methyltransferase 1 and DNA methylation patterning contribute to germinal center B-cell differentiation. Blood. 2011;118:3559–3569. - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

