Back to the origin: reconsidering replication, transcription, epigenetics, and cell cycle control
- PMID: 23634256
- PMCID: PMC3636748
- DOI: 10.1177/1947601912474891
Back to the origin: reconsidering replication, transcription, epigenetics, and cell cycle control
Abstract
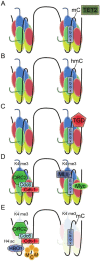
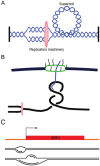
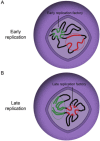
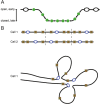
In bacteria, replication is a carefully orchestrated event that unfolds the same way for each bacterium and each cell division. The process of DNA replication in bacteria optimizes cell growth and coordinates high levels of simultaneous replication and transcription. In metazoans, the organization of replication is more enigmatic. The lack of a specific sequence that defines origins of replication has, until recently, severely limited our ability to define the organizing principles of DNA replication. This question is of particular importance as emerging data suggest that replication stress is an important contributor to inherited genetic damage and the genomic instability in tumors. We consider here the replication program in several different organisms including recent genome-wide analyses of replication origins in humans. We review recent studies on the role of cytosine methylation in replication origins, the role of transcriptional looping and gene gating in DNA replication, and the role of chromatin's 3-dimensional structure in DNA replication. We use these new findings to consider several questions surrounding DNA replication in metazoans: How are origins selected? What is the relationship between replication and transcription? How do checkpoints inhibit origin firing? Why are there early and late firing origins? We then discuss whether oncogenes promote cancer through a role in DNA replication and whether errors in DNA replication are important contributors to the genomic alterations and gene fusion events observed in cancer. We conclude with some important areas for future experimentation.
Keywords: checkpoints; epigenetics; origin; replication; replicon; transcription.
Conflict of interest statement
Figures
Similar articles
-
Replication origins and timing of temporal replication in budding yeast: how to solve the conundrum?Curr Genomics. 2010 May;11(3):199-211. doi: 10.2174/138920210791110942. Curr Genomics. 2010. PMID: 21037857 Free PMC article.
-
Genome-wide estimation of firing efficiencies of origins of DNA replication from time-course copy number variation data.BMC Bioinformatics. 2010 May 13;11:247. doi: 10.1186/1471-2105-11-247. BMC Bioinformatics. 2010. PMID: 20462459 Free PMC article.
-
Open chromatin encoded in DNA sequence is the signature of 'master' replication origins in human cells.Nucleic Acids Res. 2009 Oct;37(18):6064-75. doi: 10.1093/nar/gkp631. Epub 2009 Aug 10. Nucleic Acids Res. 2009. PMID: 19671527 Free PMC article.
-
Time to be versatile: regulation of the replication timing program in budding yeast.J Mol Biol. 2013 Nov 29;425(23):4696-705. doi: 10.1016/j.jmb.2013.09.020. Epub 2013 Sep 25. J Mol Biol. 2013. PMID: 24076190 Review.
-
Behavior of replication origins in Eukaryota - spatio-temporal dynamics of licensing and firing.Cell Cycle. 2015;14(14):2251-64. doi: 10.1080/15384101.2015.1056421. Epub 2015 Jun 1. Cell Cycle. 2015. PMID: 26030591 Free PMC article. Review.
Cited by
-
Replication Termination: Containing Fork Fusion-Mediated Pathologies in Escherichia coli.Genes (Basel). 2016 Jul 25;7(8):40. doi: 10.3390/genes7080040. Genes (Basel). 2016. PMID: 27463728 Free PMC article. Review.
-
Identification of Multiple Proteins Coupling Transcriptional Gene Silencing to Genome Stability in Arabidopsis thaliana.PLoS Genet. 2016 Jun 2;12(6):e1006092. doi: 10.1371/journal.pgen.1006092. eCollection 2016 Jun. PLoS Genet. 2016. PMID: 27253878 Free PMC article.
-
Shaping the landscape of the Escherichia coli chromosome: replication-transcription encounters in cells with an ectopic replication origin.Nucleic Acids Res. 2015 Sep 18;43(16):7865-77. doi: 10.1093/nar/gkv704. Epub 2015 Jul 8. Nucleic Acids Res. 2015. PMID: 26160884 Free PMC article.
-
Too Much of a Good Thing: How Ectopic DNA Replication Affects Bacterial Replication Dynamics.Front Microbiol. 2020 Apr 15;11:534. doi: 10.3389/fmicb.2020.00534. eCollection 2020. Front Microbiol. 2020. PMID: 32351461 Free PMC article. Review.
-
MYC and the control of DNA replication.Cold Spring Harb Perspect Med. 2014 Jun 2;4(6):a014423. doi: 10.1101/cshperspect.a014423. Cold Spring Harb Perspect Med. 2014. PMID: 24890833 Free PMC article. Review.
References
-
- Jacob F, Brenner S. [On the regulation of DNA synthesis in bacteria: the hypothesis of the replicon]. C R Hebd Seances Acad Sci. 1963;256:298-300 - PubMed
-
- Mechali M. Eukaryotic DNA replication origins: many choices for appropriate answers. Nat Rev Mol Cell Biol. 2010;11:728-38 - PubMed
-
- Bell SP, Dutta A. DNA replication in eukaryotic cells. Annu Rev Biochem. 2002;71:333-74 - PubMed
-
- Todorovic V, Falaschi A, Giacca M. Replication origins of mammalian chromosomes: the happy few. Front Biosci. 1999;4:D859-68 - PubMed
-
- Kohzaki H, Murakami Y. Transcription factors and DNA replication origin selection. Bioessays. 2005;27:1107-16 - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
